3L8R
 
 | | The crystal structure of PtcA from S. mutans | | Descriptor: | Putative PTS system, cellobiose-specific IIA component | | Authors: | Lei, J, Liu, X, Li, L. | | Deposit date: | 2010-01-03 | | Release date: | 2010-01-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structure of PtcA from Streptococcus mutans
To be Published
|
|
9IJE
 
 | | Epinephrine-activated human beta3 adrenergic receptor | | Descriptor: | Beta-3 adrenergic receptor, Camelid antibody VHH fragment, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Zheng, S, Zhang, S, Dai, S, Chen, K, Gao, K, Lin, B, Liu, X. | | Deposit date: | 2024-06-22 | | Release date: | 2024-08-21 | | Last modified: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (2.34 Å) | | Cite: | Molecular Mechanism of the beta 3 AR Agonist Activity of a beta-Blocker.
Chempluschem, 89, 2024
|
|
9IJD
 
 | | Carazolol-activated human beta3 adrenergic receptor | | Descriptor: | (2S)-1-(9H-Carbazol-4-yloxy)-3-(isopropylamino)propan-2-ol, Beta-3 adrenergic receptor, Camelid antibody VHH fragment, ... | | Authors: | Zheng, S, Zhang, S, Dai, S, Chen, K, Gao, K, Lin, B, Liu, X. | | Deposit date: | 2024-06-22 | | Release date: | 2024-08-21 | | Last modified: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | Molecular Mechanism of the beta 3 AR Agonist Activity of a beta-Blocker.
Chempluschem, 89, 2024
|
|
2F7Z
 
 | | Protein Kinase A bound to (R)-1-(1H-Indol-3-ylmethyl)-2-(2-pyridin-4-yl-[1,7]naphtyridin-5-yloxy)-ehylamine | | Descriptor: | (1S)-1-(1H-INDOL-3-YLMETHYL)-2-(2-PYRIDIN-4-YL-[1,7]NAPHTYRIDIN-5-YLOXY)-EHYLAMINE, PKI, inhibitory peptide, ... | | Authors: | Li, Q, Woods, K.W, Thomas, S, Zhu, G.D, Packard, G, Fisher, J, Li, T, Gong, J, Dinges, J, Song, X, Abrams, J, Luo, Y, Johnson, E.F, Shi, Y, Liu, X, Klinghofer, V, Des Jong, R, Oltersdorf, T, Stoll, V.S, Jakob, C.G, Rosenberg, S.H, Giranda, V.L. | | Deposit date: | 2005-12-01 | | Release date: | 2006-06-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Synthesis and structure-activity relationship of 3,4'-bispyridinylethylenes: discovery of a potent 3-isoquinolinylpyridine inhibitor of protein kinase B (PKB/Akt) for the treatment of cancer.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
3WFV
 
 | | HIV-1 CRF07 gp41 | | Descriptor: | Envelope glycoprotein gp160 | | Authors: | Du, J, Xue, H, Ma, J, Liu, F, Zhou, J, Shao, Y, Qiao, W, Liu, X. | | Deposit date: | 2013-07-24 | | Release date: | 2013-10-16 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The crystal structure of HIV CRF07 B'/C gp41 reveals a hyper-mutant site in the middle of HR2 heptad repeat
Virology, 446, 2013
|
|
3EXR
 
 | | Crystal structure of KGPDC from Streptococcus mutans | | Descriptor: | RmpD (Hexulose-6-phosphate synthase) | | Authors: | Li, G.L, Liu, X, Li, L.F, Su, X.D. | | Deposit date: | 2008-10-16 | | Release date: | 2009-08-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Open-closed conformational change revealed by the crystal structures of 3-keto-L-gulonate 6-phosphate decarboxylase from Streptococcus mutans
Biochem.Biophys.Res.Commun., 381, 2009
|
|
3EXS
 
 | | Crystal structure of KGPDC from Streptococcus mutans in complex with D-R5P | | Descriptor: | RIBULOSE-5-PHOSPHATE, RmpD (Hexulose-6-phosphate synthase) | | Authors: | Li, G.L, Liu, X, Wang, K.T, Li, L.F, Su, X.D. | | Deposit date: | 2008-10-17 | | Release date: | 2009-08-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Open-closed conformational change revealed by the crystal structures of 3-keto-L-gulonate 6-phosphate decarboxylase from Streptococcus mutans
Biochem.Biophys.Res.Commun., 381, 2009
|
|
6M03
 
 | | The crystal structure of COVID-19 main protease in apo form | | Descriptor: | 3C-like proteinase | | Authors: | Zhang, B, Zhao, Y, Jin, Z, Liu, X, Yang, H, Rao, Z. | | Deposit date: | 2020-02-19 | | Release date: | 2020-03-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for replicase polyprotein cleavage and substrate specificity of main protease from SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
8DCP
 
 | | PI 3-kinase alpha with nanobody 3-126 | | Descriptor: | Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Hart, J.R, Liu, X, Pan, C, Liang, A, Ueno, L, Xu, Y, Quezada, A, Zou, X, Yang, S, Zhou, Q, Schoonooghe, S, Hassanzadeh-Ghassabeh, G, Xia, T, Shui, W, Yang, D, Vogt, P.K, Wang, M.-W. | | Deposit date: | 2022-06-17 | | Release date: | 2022-09-21 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.41 Å) | | Cite: | Nanobodies and chemical cross-links advance the structural and functional analysis of PI3K alpha.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
8DD8
 
 | | PI 3-kinase alpha with nanobody 3-142, crosslinked with DSG | | Descriptor: | Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Hart, J.R, Liu, X, Pan, C, Liang, A, Ueno, L, Xu, Y, Quezada, A, Zou, X, Yang, S, Zhou, Q, Schoonooghe, S, Hassanzadeh-Ghassabeh, G, Xia, T, Shui, W, Yang, D, Vogt, P.K, Wang, M.-W. | | Deposit date: | 2022-06-17 | | Release date: | 2022-09-21 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Nanobodies and chemical cross-links advance the structural and functional analysis of PI3K alpha.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
8DD4
 
 | | PI 3-kinase alpha with nanobody 3-142 | | Descriptor: | Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Hart, J.R, Liu, X, Pan, C, Liang, A, Ueno, L, Xu, Y, Quezada, A, Zou, X, Yang, S, Zhou, Q, Schoonooghe, S, Hassanzadeh-Ghassabeh, G, Xia, T, Shui, W, Yang, D, Vogt, P.K, Wang, M.-W. | | Deposit date: | 2022-06-17 | | Release date: | 2022-09-21 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Nanobodies and chemical cross-links advance the structural and functional analysis of PI3K alpha.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
8DCX
 
 | | PI 3-kinase alpha with nanobody 3-159 | | Descriptor: | Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Hart, J.R, Liu, X, Pan, C, Liang, A, Ueno, L, Xu, Y, Quezada, A, Zou, X, Yang, S, Zhou, Q, Schoonooghe, S, Hassanzadeh-Ghassabeh, G, Xia, T, Shui, W, Yang, D, Vogt, P.K, Wang, M.-W. | | Deposit date: | 2022-06-17 | | Release date: | 2022-09-21 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Nanobodies and chemical cross-links advance the structural and functional analysis of PI3K alpha.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
4EQA
 
 | | Crystal structure of PA1844 in complex with PA1845 from Pseudomonas aeruginosa PAO1 | | Descriptor: | Putative uncharacterized protein | | Authors: | Shang, G, Li, N, Zhang, J, Lu, D, Yu, Q, Zhao, Y, Liu, X, Xu, S, Gu, L. | | Deposit date: | 2012-04-18 | | Release date: | 2012-09-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural insight into how Pseudomonas aeruginosa peptidoglycanhydrolase Tse1 and its immunity protein Tsi1 function.
Biochem.J., 448, 2012
|
|
5EU8
 
 | | Structure of FIPV main protease in complex with dual inhibitors | | Descriptor: | 1,2-ETHANEDIOL, N-[(5-METHYLISOXAZOL-3-YL)CARBONYL]ALANYL-L-VALYL-N~1~-((1R,2Z)-4-(BENZYLOXY)-4-OXO-1-{[(3R)-2-OXOPYRROLIDIN-3-YL]METHYL}BUT-2-ENYL)-L-LEUCINAMIDE, ZINC ION, ... | | Authors: | Wang, F, Chen, C, Liu, X, Yang, K, Xu, X, Yang, H. | | Deposit date: | 2015-11-18 | | Release date: | 2015-12-30 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.447 Å) | | Cite: | Crystal Structure of Feline Infectious Peritonitis Virus Main Protease in Complex with Synergetic Dual Inhibitors
J.Virol., 90, 2015
|
|
1Q2R
 
 | | Chemical trapping and crystal structure of a catalytic tRNA guanine transglycosylase covalent intermediate | | Descriptor: | 9-DEAZAGUANINE, Queuine tRNA-ribosyltransferase, RNA (5'-R(*AP*GP*CP*AP*CP*GP*GP*CP*UP*(N)P*UP*AP*AP*AP*CP*CP*GP*UP*GP*C)-3'), ... | | Authors: | Xie, W, Liu, X, Huang, R.H. | | Deposit date: | 2003-07-25 | | Release date: | 2003-09-09 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Chemical trapping and crystal structure of a catalytic tRNA guanine transglycosylase covalent intermediate
Nat.Struct.Biol., 10, 2003
|
|
1PUY
 
 | | 1.5 A resolution structure of a synthetic DNA hairpin with a stilbenediether linker | | Descriptor: | 5'-D(*GP*TP*TP*TP*TP*GP*(S02)P*CP*AP*AP*AP*AP*C)-3', MAGNESIUM ION | | Authors: | Egli, M, Tereshko, V, Murshudov, G, Sanishvili, R, Liu, X, Lewis, F.D. | | Deposit date: | 2003-06-25 | | Release date: | 2003-10-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Face-to-face and edge-to-face pi-pi interactions in a synthetic DNA hairpin with a stilbenediether linker
J.Am.Chem.Soc., 125, 2003
|
|
1Q2S
 
 | | Chemical trapping and crystal structure of a catalytic tRNA guanine transglycosylase covalent intermediate | | Descriptor: | 5'-R(*AP*GP*CP*AP*CP*GP*GP*CP*UP*(PQ1)P*UP*AP*AP*AP*CP*CP*GP*UP*GP*C)-3', 9-DEAZAGUANINE, Queuine tRNA-ribosyltransferase, ... | | Authors: | Xie, W, Liu, X, Huang, R.H. | | Deposit date: | 2003-07-25 | | Release date: | 2003-09-09 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Chemical trapping and crystal structure of a catalytic tRNA guanine transglycosylase covalent intermediate
Nat.Struct.Biol., 10, 2003
|
|
4ANJ
 
 | | MYOSIN VI (MDinsert2-GFP fusion) PRE-POWERSTROKE STATE (MG.ADP.AlF4) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CALCIUM ION, CALMODULIN, ... | | Authors: | Menetrey, J, Isabet, T, Ropars, V, Mukherjea, M, Pylypenko, O, Liu, X, Perez, J, Vachette, P, Sweeney, H.L, Houdusse, A.M. | | Deposit date: | 2012-03-19 | | Release date: | 2012-10-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Processive Steps in the Reverse Direction Require Uncoupling of the Lead Head Lever Arm of Myosin Vi.
Mol.Cell, 48, 2012
|
|
8IQ6
 
 | | Cryo-EM structure of Latanoprost-bound prostaglandin-F2-alpha receptor-miniGq-Nb35 complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Lv, X, Gao, K, Nie, J, Zhang, X, Zhang, S, Ren, Y, Li, Q, Huang, J, Liu, L, Zhang, X, Sun, X, Zhang, W, Liu, X. | | Deposit date: | 2023-03-15 | | Release date: | 2024-01-31 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structures of human prostaglandin F 2 alpha receptor reveal the mechanism of ligand and G protein selectivity.
Nat Commun, 14, 2023
|
|
8IQ4
 
 | | Cryo-EM structure of Carboprost-bound prostaglandin-F2-alpha receptor-miniGq-Nb35 complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Lv, X, Gao, K, Nie, J, Zhang, X, Zhang, S, Ren, Y, Li, Q, Huang, J, Liu, L, Zhang, X, Sun, X, Zhang, W, Liu, X. | | Deposit date: | 2023-03-15 | | Release date: | 2024-01-31 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structures of human prostaglandin F 2 alpha receptor reveal the mechanism of ligand and G protein selectivity.
Nat Commun, 14, 2023
|
|
2ZBB
 
 | | P43 crystal of DctBp | | Descriptor: | C4-dicarboxylate transport sensor protein dctB, MALONIC ACID | | Authors: | Zhou, Y.F, Nan, B.Y, Liu, X, Nan, J, Liang, Y.H, Panjikar, S, Ma, Q.J, Wang, Y.P, Su, X.-D. | | Deposit date: | 2007-10-18 | | Release date: | 2008-11-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | crystal structures of sensory histidine kinase DctBp
to be published
|
|
4EQ8
 
 | | Crystal structure of PA1844 from Pseudomonas aeruginosa PAO1 | | Descriptor: | GLYCEROL, Putative uncharacterized protein | | Authors: | Shang, G, Li, N, Zhang, J, Lu, D, Yu, Q, Zhao, Y, Liu, X, Xu, S, Gu, L. | | Deposit date: | 2012-04-18 | | Release date: | 2012-09-12 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.392 Å) | | Cite: | Structural insight into how Pseudomonas aeruginosa peptidoglycanhydrolase Tse1 and its immunity protein Tsi1 function.
Biochem.J., 448, 2012
|
|
5H4R
 
 | | the complex of Glycoside Hydrolase 5 Lichenase from Caldicellulosiruptor sp. F32 E188Q mutant and cellotetraose | | Descriptor: | Beta-1,3-1,4-glucanase, GLYCEROL, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Dong, S, Zhou, H, Liu, X, Wang, X, Feng, Y. | | Deposit date: | 2016-11-02 | | Release date: | 2017-09-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.70310438 Å) | | Cite: | Structural insights into the substrate specificity of a glycoside hydrolase family 5 lichenase from Caldicellulosiruptor sp. F32
Biochem. J., 474, 2017
|
|
5BJS
 
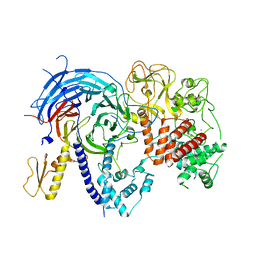 | | Apo ctPRC2 in an autoinhibited state | | Descriptor: | Histone-lysine N-methyltransferase EZH2, Polycomb protein SUZ12, Polycomb Protein EED, ... | | Authors: | Bratkowski, M.A, Liu, X. | | Deposit date: | 2016-10-22 | | Release date: | 2017-06-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.189 Å) | | Cite: | Polycomb repressive complex 2 in an autoinhibited state.
J. Biol. Chem., 292, 2017
|
|
3CXP
 
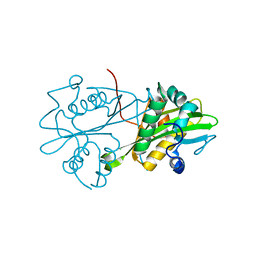 | | Crystal structure of human glucosamine 6-phosphate N-acetyltransferase 1 mutant E156A | | Descriptor: | CHLORIDE ION, Glucosamine 6-phosphate N-acetyltransferase | | Authors: | Wang, J, Liu, X, Li, L.-F, Su, X.-D. | | Deposit date: | 2008-04-25 | | Release date: | 2008-09-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Acceptor substrate binding revealed by crystal structure of human glucosamine-6-phosphate N-acetyltransferase 1
Febs Lett., 582, 2008
|
|
