4G1R
 
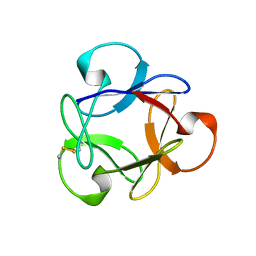 | | Crystal structure of anti-HIV actinohivin in complex with alphs-1,2-mannobiose (Form II) | | Descriptor: | Actinohivin, alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose | | Authors: | Hoque, M.M, Suzuki, K, Tsunoda, M, Jiang, J, Zhang, F, Takahashi, A, Naomi, O, Zhang, X, Sekiguchi, T, Tanaka, H, Omura, S, Takenaka, A. | | Deposit date: | 2012-07-11 | | Release date: | 2013-07-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Matured structure of anti-HIV lectin actinohivin in complex with alpha-1,2-mannobiose
To be Published
|
|
1AHK
 
 | | DER F 2, THE MAJOR MITE ALLERGEN FROM DERMATOPHAGOIDES FARINAE, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | DER F 2 | | Authors: | Ichikawa, S, Hatanaka, H, Yuuki, T, Iwamoto, N, Ogura, K, Okumura, Y, Inagaki, F. | | Deposit date: | 1997-04-07 | | Release date: | 1998-04-08 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of Der f 2, the major mite allergen for atopic diseases.
J.Biol.Chem., 273, 1998
|
|
6KV0
 
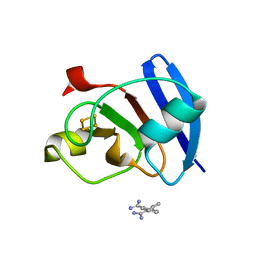 | | Ferredoxin I from C. reinhardtii, high X-ray dose | | Descriptor: | BENZAMIDINE, CHLORIDE ION, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Onishi, Y, Kurisu, G, Tanaka, H. | | Deposit date: | 2019-09-03 | | Release date: | 2020-05-20 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | X-ray dose-dependent structural changes of the [2Fe-2S] ferredoxin from Chlamydomonas reinhardtii.
J.Biochem., 167, 2020
|
|
6LK1
 
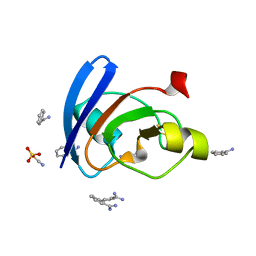 | | Ultrahigh resolution X-ray structure of Ferredoxin I from C. reinhardtii | | Descriptor: | BENZAMIDINE, FE2/S2 (INORGANIC) CLUSTER, Ferredoxin, ... | | Authors: | Onishi, Y, Kurisu, G, Tanaka, H. | | Deposit date: | 2019-12-17 | | Release date: | 2020-05-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (0.9 Å) | | Cite: | X-ray dose-dependent structural changes of the [2Fe-2S] ferredoxin from Chlamydomonas reinhardtii.
J.Biochem., 167, 2020
|
|
6KUM
 
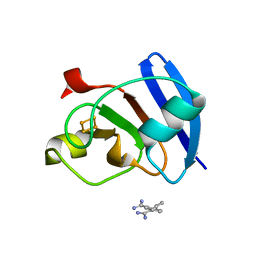 | | Ferredoxin I from C. reinhardtii, low X-ray dose | | Descriptor: | BENZAMIDINE, CHLORIDE ION, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Onishi, Y, Kurisu, G, Tanaka, H. | | Deposit date: | 2019-09-02 | | Release date: | 2020-05-20 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | X-ray dose-dependent structural changes of the [2Fe-2S] ferredoxin from Chlamydomonas reinhardtii.
J.Biochem., 167, 2020
|
|
4HLN
 
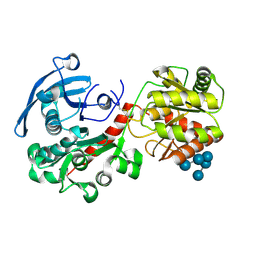 | | Structure of barley starch synthase I in complex with maltooligosaccharide | | Descriptor: | Starch synthase I, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Cuesta-Seijo, J.A, Nielsen, M.M, Marri, L, Tanaka, H, Palcic, M.M. | | Deposit date: | 2012-10-17 | | Release date: | 2013-06-05 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of starch synthase I from barley: insight into regulatory mechanisms of starch synthase activity
Acta Crystallogr.,Sect.D, 69, 2013
|
|
1BQ5
 
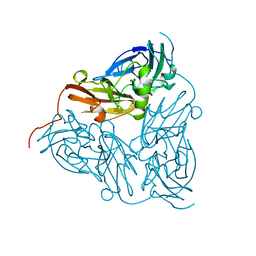 | | NITRITE REDUCTASE FROM ALCALIGENES XYLOSOXIDANS GIFU 1051 | | Descriptor: | COPPER (II) ION, NITRITE REDUCTASE | | Authors: | Inoue, T, Gotowda, M, Deligeer, Suzuki, S, Kataoka, K, Yamaguchi, K, Watanabe, H, Goho, M, Yasushi, K.A.I. | | Deposit date: | 1998-08-21 | | Release date: | 1999-08-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Type 1 Cu structure of blue nitrite reductase from Alcaligenes xylosoxidans GIFU 1051 at 2.05 A resolution: comparison of blue and green nitrite reductases.
J.Biochem.(Tokyo), 124, 1998
|
|
2E0Z
 
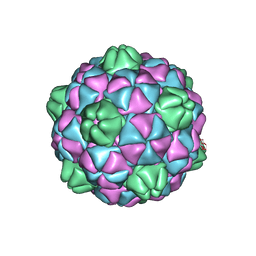 | | Crystal structure of virus-like particle from Pyrococcus furiosus | | Descriptor: | Virus-like particle | | Authors: | Akita, F, Chong, K.T, Tanaka, H, Yamashita, E, Miyazaki, N, Nakaishi, Y, Namba, K, Ono, Y, Suzuki, M, Tsukihara, T, Nakagawa, A. | | Deposit date: | 2006-10-16 | | Release date: | 2007-04-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | The Crystal Structure of a Virus-like Particle from the Hyperthermophilic Archaeon Pyrococcus furiosus Provides Insight into the Evolution of Viruses
J.Mol.Biol., 368, 2007
|
|
6J13
 
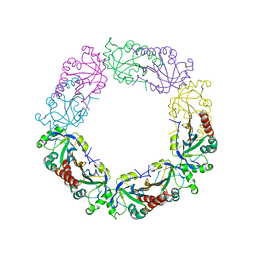 | | Redox protein from Chlamydomonas reinhardtii | | Descriptor: | 2-cys peroxiredoxin | | Authors: | Charoenwattansatien, R, Zinzius, K, Tanaka, H, Hippler, M, Kurisu, G. | | Deposit date: | 2018-12-27 | | Release date: | 2019-12-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Calcium sensing via EF-hand 4 enables thioredoxin activity in the sensor-responder protein calredoxin in the green algaChlamydomonas reinhardtii.
J.Biol.Chem., 295, 2020
|
|
3SNL
 
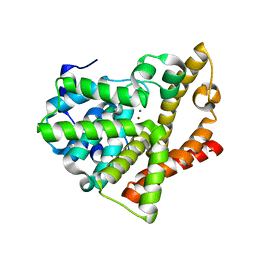 | | Highly Potent, Selective, and Orally Active Phosphodiestarase 10A Inhibitors | | Descriptor: | 6-chloro-3,4-dimethyl-1-(3-methylpyridin-4-yl)-8-(trifluoromethyl)imidazo[1,5-a]quinoxaline, MAGNESIUM ION, ZINC ION, ... | | Authors: | Malamas, M.S, Ni, Y, Erdei, J, Stange, H, Schindler, R, Lankau, H.-J, Grunwald, C, Fan, K.Y, Parris, K.D, Langen, B, Egerland, U, Hage, T, Marquis, K.L, Grauer, S, Brennan, J, Navarra, R, Graf, R, Harrison, B.L, Robichaud, A, Kronbach, T, Pangalos, M, Hofgen, N, Brandon, N.J. | | Deposit date: | 2011-06-29 | | Release date: | 2011-10-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Highly Potent, Selective, and Orally Active Phosphodiesterase 10A Inhibitors.
J.Med.Chem., 54, 2011
|
|
3WIQ
 
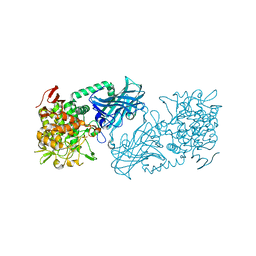 | | Crystal structure of kojibiose phosphorylase complexed with kojibiose | | Descriptor: | Kojibiose phosphorylase, SULFATE ION, alpha-D-glucopyranose-(1-2)-beta-D-glucopyranose | | Authors: | Okada, S, Yamamoto, T, Watanabe, H, Nishimoto, T, Chaen, H, Fukuda, S, Wakagi, T, Fushinobu, S. | | Deposit date: | 2013-09-24 | | Release date: | 2014-02-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural and mutational analysis of substrate recognition in kojibiose phosphorylase
Febs J., 281, 2014
|
|
3SNI
 
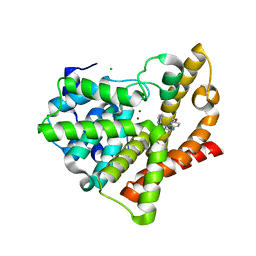 | | Highly Potent, Selective, and Orally Active Phosphodiestarase 10A Inhibitors | | Descriptor: | 2-methoxy-6,7-dimethyl-9-(4-methylpyridin-3-yl)imidazo[1,5-a]pyrido[3,2-e]pyrazine, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Malamas, M.S, Ni, Y, Erdei, J, Stange, H, Schindler, R, Lankau, H.-J, Grunwald, C, Fan, K.Y, Parris, K.D, Langen, B, Egerland, U, Hage, T, Marquis, K.L, Grauer, S, Brennan, J, Navarra, R, Graf, R, Harrison, B.L, Robichaud, A, Kronbach, T, Pangalos, M, Hofgen, N, Brandon, N.J. | | Deposit date: | 2011-06-29 | | Release date: | 2011-10-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Highly Potent, Selective, and Orally Active Phosphodiesterase 10A Inhibitors.
J.Med.Chem., 54, 2011
|
|
3WIR
 
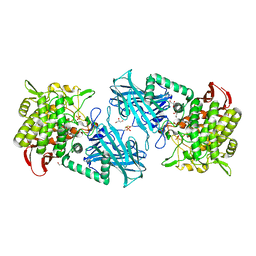 | | Crystal structure of kojibiose phosphorylase complexed with glucose | | Descriptor: | GLYCEROL, Kojibiose phosphorylase, PHOSPHATE ION, ... | | Authors: | Okada, S, Yamamoto, T, Watanabe, H, Nishimoto, T, Chaen, H, Fukuda, S, Wakagi, T, Fushinobu, S. | | Deposit date: | 2013-09-24 | | Release date: | 2014-02-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural and mutational analysis of substrate recognition in kojibiose phosphorylase
Febs J., 281, 2014
|
|
7X7O
 
 | | SARS-CoV-2 spike RBD in complex with neutralizing antibody UT28K | | Descriptor: | Spike protein S1, UT28K Fab, heavy chain, ... | | Authors: | Ozawa, T, Tani, H, Anraku, Y, Kita, S, Igarashi, E, Saga, Y, Inasaki, N, Kawasuji, H, Yamada, H, Sasaki, S, Somekawa, M, Sasaki, J, Hayakawa, Y, Yamamoto, Y, Morinaga, Y, Kurosawa, N, Isobe, M, Fukuhara, H, Maenaka, K, Hashiguchi, T, Kishi, H, Kitajima, I, Saito, S, Niimi, H. | | Deposit date: | 2022-03-10 | | Release date: | 2022-05-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.75 Å) | | Cite: | Novel super-neutralizing antibody UT28K is capable of protecting against infection from a wide variety of SARS-CoV-2 variants.
Mabs, 14, 2022
|
|
2V5E
 
 | | The structure of the GDNF:Coreceptor complex: Insights into RET signalling and heparin binding. | | Descriptor: | 1,2-ETHANEDIOL, 1,3,4,6-tetra-O-sulfo-beta-D-fructofuranose-(2-1)-2,3,4,6-tetra-O-sulfonato-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Parkash, V, Leppanen, V.-M, Virtanen, H, Jurvansuu, J.-M, Bespalov, M.M, Sidorova, Y.A, Runeberg-Roos, P, Saarma, M, Goldman, A. | | Deposit date: | 2008-10-03 | | Release date: | 2008-10-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The Structure of the Glial Cell Line-Derived Neurotrophic Factor-Coreceptor Complex: Insights Into Ret Signaling and Heparin Binding.
J.Biol.Chem., 283, 2008
|
|
4END
 
 | | Crystal structure of anti-HIV actinohivin in complex with alpha-1,2-mannobiose (P 2 21 21 form) | | Descriptor: | ACETONITRILE, Actinohivin, alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose | | Authors: | Hoque, M.M, Suzuki, K, Tsunoda, M, Jiang, J, Zhang, F, Takahashi, A, Naomi, O, Zhang, X, Sekiguchi, T, Tanaka, H, Omura, S, Takenaka, A. | | Deposit date: | 2012-04-13 | | Release date: | 2013-07-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Matured structure of anti-HIV lectin actinohivin in complex with 1,2-mannobiose
To be Published
|
|
4DEN
 
 | | Structural insightsinto potent, specific anti-HIV property of actinohivin; Crystal structure of actinohivin in complex with alpha(1-2) mannobiose moiety of high-mannose type glycan of gp120 | | Descriptor: | Actinohivin, POTASSIUM ION, alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose | | Authors: | Hoque, M.M, Suzuki, K, Tsunoda, M, Jiang, J, Zhang, F, Takahashi, A, Naomi, O, Zhang, X, Sekiguchi, T, Tanaka, H, Omura, S, Takenaka, A. | | Deposit date: | 2012-01-20 | | Release date: | 2012-11-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural insights into the specific anti-HIV property of actinohivin: structure of its complex with the alpha(1–2)mannobiose moiety of gp120
Acta Crystallogr.,Sect.D, 68, 2012
|
|
1MKN
 
 | | N-TERMINAL HALF OF MIDKINE | | Descriptor: | PROTEIN (MIDKINE) | | Authors: | Iwasaki, W, Nagata, K, Hatanaka, H, Ogura, K, Inui, T, Kimura, T, Muramatsu, T, Yoshida, K, Tasumi, M, Inagaki, F. | | Deposit date: | 1999-03-16 | | Release date: | 1999-03-23 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of midkine, a new heparin-binding growth factor.
EMBO J., 16, 1997
|
|
8IFJ
 
 | |
1J03
 
 | | Solution structure of a putative steroid-binding protein from Arabidopsis | | Descriptor: | putative steroid binding protein | | Authors: | Suzuki, S, Hatanaka, H, Kigawa, T, Terada, T, Shirouzu, M, Seki, M, Shinozaki, K, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-10-29 | | Release date: | 2003-12-16 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of an Arabidopsis homologue of the mammalian membrane-associated progesterone receptor
To be Published
|
|
1WOT
 
 | | Structure of putative minimal nucleotidyltransferase | | Descriptor: | PUTATIVE MINIMAL NUCLEOTIDYLTRANSFERASE | | Authors: | Suzuki, S, Hatanaka, H, Hondoh, T, Okumura, A, Kuroda, Y, Kuramitsu, S, Shibata, T, Inoue, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-08-25 | | Release date: | 2004-09-07 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure of Putative Minimal Nucleotidyltransferase
To be Published
|
|
7VW6
 
 | | Cryo-EM Structure of Formate Dehydrogenase 1 from Methylorubrum extorquens AM1 | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, FE2/S2 (INORGANIC) CLUSTER, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Yoshikawa, T, Makino, F, Miyata, T, Suzuki, Y, Tanaka, H, Namba, K, Sowa, K, Kitazumi, Y, Shirai, O. | | Deposit date: | 2021-11-09 | | Release date: | 2022-06-01 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (2.19 Å) | | Cite: | Multiple electron transfer pathways of tungsten-containing formate dehydrogenase in direct electron transfer-type bioelectrocatalysis.
Chem.Commun.(Camb.), 58, 2022
|
|
1X2H
 
 | | Crystal Structure of Lipate-Protein Ligase A from Escherichia coli complexed with lipoic acid | | Descriptor: | LIPOIC ACID, Lipoate-protein ligase A | | Authors: | Fujiwara, K, Toma, S, Okamura-Ikeda, K, Motokawa, Y, Nakagawa, A, Taniguchi, H. | | Deposit date: | 2005-04-23 | | Release date: | 2005-08-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Crystal structure of lipoate-protein ligase A from Escherichia coli: Determination of the lipoic acid-binding site
J.Biol.Chem., 280, 2005
|
|
1X2G
 
 | | Crystal Structure of Lipate-Protein Ligase A from Escherichia coli | | Descriptor: | Lipoate-protein ligase A | | Authors: | Fujiwara, K, Toma, S, Okamura-Ikeda, K, Motokawa, Y, Nakagawa, A, Taniguchi, H. | | Deposit date: | 2005-04-23 | | Release date: | 2005-08-02 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of lipoate-protein ligase A from Escherichia coli: Determination of the lipoic acid-binding site
J.Biol.Chem., 280, 2005
|
|
1TV0
 
 | | Solution structure of cryptdin-4, the most potent alpha-defensin from mouse Paneth cells | | Descriptor: | Cryptdin-4 | | Authors: | Jing, W, Hunter, H.N, Tanabe, H, Ouellette, A.J, Vogel, H.J. | | Deposit date: | 2004-06-25 | | Release date: | 2005-01-04 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Cryptdin-4, a Mouse Paneth Cell alpha-Defensin.
Biochemistry, 43, 2004
|
|
