8ULH
 
 | |
8ULJ
 
 | |
7JHX
 
 | | Crystal structure of hEPG5 LIR/GABARAPL1 complex | | Descriptor: | Ectopic P granules protein 5 homolog, Gamma-aminobutyric acid receptor-associated protein-like 1, SULFATE ION | | Authors: | Cheung, Y.W.S, Yip, C.K. | | Deposit date: | 2020-07-21 | | Release date: | 2021-03-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Insights on autophagosome-lysosome tethering from structural and biochemical characterization of human autophagy factor EPG5.
Commun Biol, 4, 2021
|
|
2WD4
 
 | | Ascorbate Peroxidase as a heme oxygenase: w41A variant product with t-butyl peroxide | | Descriptor: | 3-[2-[[3-(2-CARBOXYETHYL)-5-[[3-ETHENYL-4-METHYL-5-[(2-METHYLPROPAN-2-YL)OXY]-1H-PYRROL-2-YL]METHYL]-4-METHYL-1H-PYRROL -2-YL]METHYL]-5-[(Z)-(4-ETHENYL-3-METHYL-5-OXO-PYRROL-2-YLIDENE)METHYL]-4-METHYL-1H-PYRROL-3-YL]PROPANOIC ACID, ASCORBATE PEROXIDASE, FE (III) ION, ... | | Authors: | Badyal, S.K, Metcalfe, C.L, Gumiero, A, Raven, E.L, Moody, P.C.E. | | Deposit date: | 2009-03-19 | | Release date: | 2009-04-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Evidence for Heme Oxygenase Activity in a Heme Peroxidase.
Biochemistry, 48, 2009
|
|
3DK5
 
 | | Crystal Structure of Apo-GlmU from Mycobacterium tuberculosis | | Descriptor: | Bifunctional protein glmU, MAGNESIUM ION | | Authors: | Verma, S.K, Prakash, B. | | Deposit date: | 2008-06-24 | | Release date: | 2009-05-19 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | PknB-mediated phosphorylation of a novel substrate, N-acetylglucosamine-1-phosphate uridyltransferase, modulates its acetyltransferase activity.
J.Mol.Biol., 386, 2009
|
|
3DNT
 
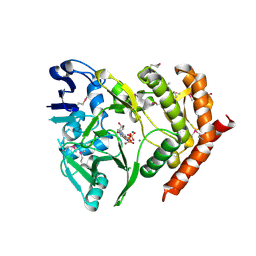 | | structures of MDT proteins | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Protein hipA, ... | | Authors: | Schumacher, M.A. | | Deposit date: | 2008-07-02 | | Release date: | 2009-01-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Molecular mechanisms of HipA-mediated multidrug tolerance and its neutralization by HipB.
Science, 323, 2009
|
|
3DNV
 
 | | MDT Protein | | Descriptor: | DNA (5'-D(*DAP*DCP*DTP*DAP*DTP*DCP*DCP*DCP*DCP*DTP*DTP*DAP*DAP*DGP*DGP*DGP*DGP*DAP*DTP*DAP*DG)-3'), HTH-type transcriptional regulator hipB, Protein hipA, ... | | Authors: | schumacher, M.A. | | Deposit date: | 2008-07-02 | | Release date: | 2009-01-27 | | Last modified: | 2023-04-05 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Molecular mechanisms of HipA-mediated multidrug tolerance and its neutralization by HipB.
Science, 323, 2009
|
|
4KHP
 
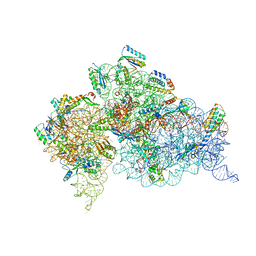 | | Structure of the Thermus thermophilus 30S ribosomal subunit in complex with de-6-MSA-pactamycin | | Descriptor: | 16S Ribosomal RNA, 30S Ribosomal protein S10, 30S Ribosomal protein S11, ... | | Authors: | Tourigny, D.S, Fernandez, I.S, Kelley, A.C, Ramakrishnan, V. | | Deposit date: | 2013-05-01 | | Release date: | 2013-06-05 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal Structure of a Bioactive Pactamycin Analog Bound to the 30S Ribosomal Subunit.
J.Mol.Biol., 425, 2013
|
|
2KQT
 
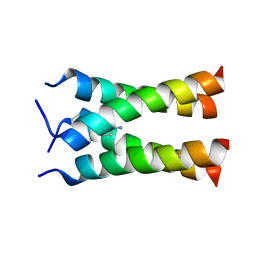 | | Solid-state NMR structure of the M2 transmembrane peptide of the influenza A virus in DMPC lipid bilayers bound to deuterated amantadine | | Descriptor: | (3S,5S,7S)-tricyclo[3.3.1.1~3,7~]decan-1-amine, M2 protein | | Authors: | Cady, S.D, Schmidt-Rohr, K, Wang, J, Soto, C.S, DeGrado, W.F, Hong, M. | | Deposit date: | 2009-11-18 | | Release date: | 2010-02-09 | | Last modified: | 2024-05-08 | | Method: | SOLID-STATE NMR | | Cite: | Structure of the amantadine binding site of influenza M2 proton channels in lipid bilayers
Nature, 463, 2010
|
|
4J47
 
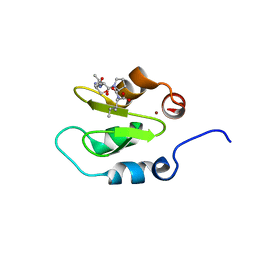 | | Crystal structure of XIAP-BIR2 domain with SVPI bound | | Descriptor: | E3 ubiquitin-protein ligase XIAP, PEPTIDE (SER-VAL-PRO-ILE), ZINC ION | | Authors: | Lukacs, C.M, Janson, C.A. | | Deposit date: | 2013-02-06 | | Release date: | 2013-09-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | The structure of XIAP BIR2: understanding the selectivity of the BIR domains.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4J3Y
 
 | | Crystal structure of XIAP-BIR2 domain | | Descriptor: | E3 ubiquitin-protein ligase XIAP, ZINC ION | | Authors: | Lukacs, C.M, Janson, C.A. | | Deposit date: | 2013-02-06 | | Release date: | 2013-09-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The structure of XIAP BIR2: understanding the selectivity of the BIR domains.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
2FSZ
 
 | |
5ZKH
 
 | |
2I4X
 
 | | HIV-1 Protease I84V, L90M with GS-8374 | | Descriptor: | DIETHYL ({4-[(2S,3R)-2-({[(3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YLOXY]CARBONYL}AMINO)-3-HYDROXY-4-{ISOBUTYL[(4-METHOXYPHENYL)SULFONYL]AMINO}BUTYL]PHENOXY}METHYL)PHOSPHONATE, Protease | | Authors: | Hatada, M. | | Deposit date: | 2006-08-22 | | Release date: | 2007-08-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Suppression of HIV-1 Protease Inhibitor Resistance by Phosphonate-mediated Solvent Anchoring.
J.Mol.Biol., 363, 2006
|
|
4Q13
 
 | |
4PXM
 
 | |
1FV2
 
 | | The Hc fragment of tetanus toxin complexed with an analogue of its ganglioside receptor GT1B | | Descriptor: | ETHYL-TRIMETHYL-SILANE, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-galactopyranose-(1-4)-[N-acetyl-alpha-neuraminic acid-(2-8)-N-acetyl-beta-neuraminic acid-(2-3)]beta-D-galactopyranose-(1-4)-beta-D-glucopyranose, PHOSPHATE ION, ... | | Authors: | Fotinou, C, Emsley, P, Black, I, Ando, H, Ishida, H, Kiso, M, Sinha, K.A, Fairweather, N.F, Isaacs, N.W. | | Deposit date: | 2000-09-18 | | Release date: | 2001-09-05 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structure of tetanus toxin Hc fragment complexed with a synthetic GT1b analogue suggests cross-linking between ganglioside receptors and the toxin.
J.Biol.Chem., 276, 2001
|
|
4Q50
 
 | |
2I4W
 
 | | HIV-1 protease WT with GS-8374 | | Descriptor: | DIETHYL ({4-[(2S,3R)-2-({[(3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YLOXY]CARBONYL}AMINO)-3-HYDROXY-4-{ISOBUTYL[(4-METHOXYPHENYL)SULFONYL]AMINO}BUTYL]PHENOXY}METHYL)PHOSPHONATE, Protease | | Authors: | Hatada, M. | | Deposit date: | 2006-08-22 | | Release date: | 2007-08-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Suppression of HIV-1 Protease Inhibitor Resistance by Phosphonate-mediated Solvent Anchoring.
J.Mol.Biol., 363, 2006
|
|
2I4D
 
 | | Crystal structure of WT HIV-1 protease with GS-8373 | | Descriptor: | ({4-[(2S,3R)-2-({[(3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YLOXY]CARBONYL}AMINO)-3-HYDROXY-4-{ISOBUTYL[(4-METHOXYPHENYL)SULFONYL]AMINO}BUTYL]PHENOXY}METHYL)PHOSPHONIC ACID, Protease | | Authors: | Hatada, M. | | Deposit date: | 2006-08-21 | | Release date: | 2007-08-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Suppression of HIV-1 Protease Inhibitor Resistance by Phosphonate-mediated Solvent Anchoring.
J.Mol.Biol., 363, 2006
|
|
2I4V
 
 | | HIV-1 protease I84V, L90M with TMC126 | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL [(1S,2R)-1-BENZYL-2-HYDROXY-3-{ISOBUTYL[(4-METHOXYPHENYL)SULFONYL]AMINO}PROPYL]CARBAMATE, Protease | | Authors: | Hatada, M. | | Deposit date: | 2006-08-22 | | Release date: | 2007-08-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Suppression of HIV-1 Protease Inhibitor Resistance by Phosphonate-mediated Solvent Anchoring.
J.Mol.Biol., 363, 2006
|
|
1CA5
 
 | | INTERCALATION SITE OF HYPERTHERMOPHILE CHROMOSOMAL PROTEIN SSO7D/SAC7D BOUND TO DNA | | Descriptor: | 5'-D(*GP*TP*GP*AP*TP*CP*AP*C)-3', CHROMOSOMAL PROTEIN SAC7D | | Authors: | Su, S, Gao, Y.-G, Robinson, H, Shriver, J.W, Wang, A.H.-J. | | Deposit date: | 1999-02-23 | | Release date: | 2000-02-23 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of the chromosomal proteins Sso7d/Sac7d bound to DNA containing T-G mismatched base-pairs
J.Mol.Biol., 303, 2000
|
|
2I4U
 
 | | HIV-1 protease with TMC-126 | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL [(1S,2R)-1-BENZYL-2-HYDROXY-3-{ISOBUTYL[(4-METHOXYPHENYL)SULFONYL]AMINO}PROPYL]CARBAMATE, Protease | | Authors: | Hatada, M. | | Deposit date: | 2006-08-22 | | Release date: | 2007-08-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Suppression of HIV-1 Protease Inhibitor Resistance by Phosphonate-mediated Solvent Anchoring.
J.Mol.Biol., 363, 2006
|
|
2H95
 
 | |
1CA6
 
 | | INTERCALATION SITE OF HYPERTHERMOPHILE CHROMOSOMAL PROTEIN SSO7D/SAC7D BOUND TO DNA | | Descriptor: | 5'-D(*GP*TP*GP*AP*TP*CP*GP*C)-3', CHROMOSOMAL PROTEIN SAC7D | | Authors: | Su, S, Gao, Y.-G, Robinson, H, Shriver, J.W, Wang, A.H.-J. | | Deposit date: | 1999-02-23 | | Release date: | 2000-02-23 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of the chromosomal proteins Sso7d/Sac7d bound to DNA containing T-G mismatched base-pairs
J.Mol.Biol., 303, 2000
|
|
