7RR0
 
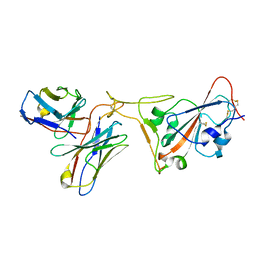 | | SARS-CoV-2 receptor binding domain bound to Fab PDI 222 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, PDI 222 Fab Heavy Chain, PDI 222 Fab Light Chain, ... | | Authors: | Pymm, P, Glukhova, A, Black, K.A, Tham, W.H. | | Deposit date: | 2021-08-08 | | Release date: | 2021-10-06 | | Last modified: | 2021-10-27 | | Method: | ELECTRON MICROSCOPY (3.12 Å) | | Cite: | Landscape of human antibody recognition of the SARS-CoV-2 receptor binding domain.
Cell Rep, 37, 2021
|
|
7QQR
 
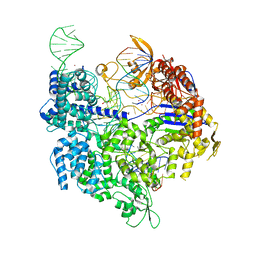 | | SpCas9 bound to AAVS1 off-target5 DNA substrate | | Descriptor: | 1,2-ETHANEDIOL, AAVS1 off-target5 non-target strand, AAVS1 off-target5 target strand, ... | | Authors: | Pacesa, M, Jinek, M. | | Deposit date: | 2022-01-10 | | Release date: | 2022-10-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural basis for Cas9 off-target activity.
Cell, 185, 2022
|
|
7QQV
 
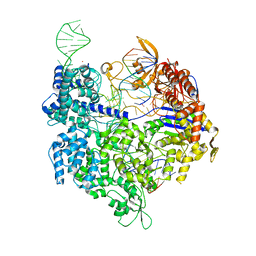 | | SpCas9 bound to FANCF off-target3 DNA substrate | | Descriptor: | CRISPR-associated endonuclease Cas9/Csn1, FANCF off-target3 non-target strand, FANCF off-target3 target strand, ... | | Authors: | Pacesa, M, Jinek, M. | | Deposit date: | 2022-01-10 | | Release date: | 2022-10-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural basis for Cas9 off-target activity.
Cell, 185, 2022
|
|
7QR5
 
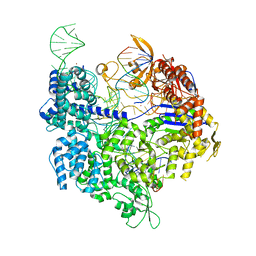 | | SpCas9 bound to FANCF off-target6 DNA substrate | | Descriptor: | 1,2-ETHANEDIOL, CRISPR-associated endonuclease Cas9/Csn1, FANCF off-target6 non-target strand, ... | | Authors: | Pacesa, M, Jinek, M. | | Deposit date: | 2022-01-17 | | Release date: | 2022-10-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural basis for Cas9 off-target activity.
Cell, 185, 2022
|
|
7QQO
 
 | | SpCas9 bound to AAVS1 off-target1 DNA substrate | | Descriptor: | AAVS1 off-target1 non-target strand, AAVS1 off-target1 target strand, AAVS1 sgRNA, ... | | Authors: | Pacesa, M, Jinek, M. | | Deposit date: | 2022-01-10 | | Release date: | 2022-10-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for Cas9 off-target activity.
Cell, 185, 2022
|
|
7QQQ
 
 | | SpCas9 bound to AAVS1 off-target4 DNA substrate | | Descriptor: | 1,2-ETHANEDIOL, AAVS1 off-target4 non-target strand, AAVS1 off-target4 target strand, ... | | Authors: | Pacesa, M, Jinek, M. | | Deposit date: | 2022-01-10 | | Release date: | 2022-10-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural basis for Cas9 off-target activity.
Cell, 185, 2022
|
|
7QQU
 
 | | SpCas9 bound to FANCF off-target2 DNA substrate | | Descriptor: | CRISPR-associated endonuclease Cas9/Csn1, FANCF off-target2 non-target strand, FANCF off-target2 target strand, ... | | Authors: | Pacesa, M, Jinek, M. | | Deposit date: | 2022-01-10 | | Release date: | 2022-10-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural basis for Cas9 off-target activity.
Cell, 185, 2022
|
|
7QQT
 
 | | SpCas9 bound to FANCF off-target1 DNA substrate | | Descriptor: | CRISPR-associated endonuclease Cas9/Csn1, FANCF off-target1 non-target strand, FANCF off-target1 target strand, ... | | Authors: | Pacesa, M, Jinek, M. | | Deposit date: | 2022-01-10 | | Release date: | 2022-10-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for Cas9 off-target activity.
Cell, 185, 2022
|
|
7QR8
 
 | | SpCas9 bound to PTPRC off-target1 DNA substrate | | Descriptor: | CRISPR-associated endonuclease Cas9/Csn1, MAGNESIUM ION, POTASSIUM ION, ... | | Authors: | Pacesa, M, Jinek, M. | | Deposit date: | 2022-01-10 | | Release date: | 2022-10-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural basis for Cas9 off-target activity.
Cell, 185, 2022
|
|
7ELR
 
 | | Crystal structure of xanthine riboswitch with xanthine | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, NMT1 (46-MER), ... | | Authors: | Xu, X.C, Ren, A.M. | | Deposit date: | 2021-04-12 | | Release date: | 2021-06-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Insights into xanthine riboswitch structure and metal ion-mediated ligand recognition.
Nucleic Acids Res., 49, 2021
|
|
7ELS
 
 | | Crystal structure of xanthine riboswitch with 8-azaxanthine | | Descriptor: | 8-AZAXANTHINE, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Xu, X.C, Ren, A.M. | | Deposit date: | 2021-04-12 | | Release date: | 2021-06-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Insights into xanthine riboswitch structure and metal ion-mediated ligand recognition.
Nucleic Acids Res., 49, 2021
|
|
7ELP
 
 | |
7ELQ
 
 | |
7Z14
 
 | | Cryo-EM structure of Torpedo nicotinic acetylcholine receptor in complex with a short-chain neurotoxin. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Acetylcholine receptor subunit alpha, ... | | Authors: | Nys, M.A.E.M, Zarkadas, E, Ulens, C, Nury, H. | | Deposit date: | 2022-02-24 | | Release date: | 2022-08-17 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | The molecular mechanism of snake short-chain alpha-neurotoxin binding to muscle-type nicotinic acetylcholine receptors.
Nat Commun, 13, 2022
|
|
4I4T
 
 | | Crystal structure of tubulin-RB3-TTL-Zampanolide complex | | Descriptor: | (2Z,4E)-N-[(S)-[(1S,2E,5S,8E,10Z,17S)-3,11-dimethyl-19-methylidene-7,13-dioxo-6,21-dioxabicyclo[15.3.1]henicosa-2,8,10-trien-5-yl](hydroxy)methyl]hexa-2,4-dienamide, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Prota, A.E, Steinmetz, M.O. | | Deposit date: | 2012-11-28 | | Release date: | 2013-01-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular Mechanism of Action of Microtubule-Stabilizing Anticancer Agents.
Science, 339, 2013
|
|
7ZX2
 
 | | Tubulin-Pelophen B complex | | Descriptor: | (3R,4S,7S,9S,11S)-3,4,11-trihydroxy-7-((R,Z)-4-(hydroxymethyl)hex-2-en-2-yl)-9-methoxy-12,12-dimethyl-6-oxa-1(1,3)-benzenacyclododecaphan-5-one, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Estevez-Gallego, J, Diaz, J.F, Van der Eycken, J, Oliva, M.A. | | Deposit date: | 2022-05-20 | | Release date: | 2022-11-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Chemical modulation of microtubule structure through the laulimalide/peloruside site.
Structure, 31, 2023
|
|
8A0L
 
 | | Tubulin-CW1-complex | | Descriptor: | (3~{S},4~{R},8~{S},10~{S},12~{S},14~{S})-14-[(~{Z},4~{R})-4-(hydroxymethyl)hex-2-en-2-yl]-4,12-dimethoxy-9,9-dimethyl-3,8,10-tris(oxidanyl)-1-oxacyclotetradecan-2-one, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Prota, A.E, Diaz, J.F, Steinmetz, M.O, Oliva, M.A. | | Deposit date: | 2022-05-28 | | Release date: | 2022-12-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9981 Å) | | Cite: | Chemical modulation of microtubule structure through the laulimalide/peloruside site.
Structure, 31, 2023
|
|
6GS4
 
 | | Crystal structure of peptide transporter DtpA-nanobody in complex with valganciclovir | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, Dipeptide and tripeptide permease A, [(2~{S})-2-[(2-azanyl-6-oxidanylidene-3~{H}-purin-9-yl)methoxy]-3-oxidanyl-propyl] (2~{S})-2-azanyl-3-methyl-butanoate, ... | | Authors: | Ural-Blimke, Y, Flayhan, A, Quistgaard, E.M, Loew, C. | | Deposit date: | 2018-06-13 | | Release date: | 2019-01-30 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.645 Å) | | Cite: | Structure of Prototypic Peptide Transporter DtpA from E. coli in Complex with Valganciclovir Provides Insights into Drug Binding of Human PepT1.
J. Am. Chem. Soc., 141, 2019
|
|
1EIQ
 
 | | 2,3-DIHYDROXYBIPHENYL-1,2-DIOXYGENASE | | Descriptor: | 2,3-DIHYDROXYBIPHENYL-1,2-DIOXYGENASE, FE (III) ION | | Authors: | Senda, T. | | Deposit date: | 2000-02-28 | | Release date: | 2001-02-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of substrate free and complex forms of reactivated BphC, an extradiol type ring-cleavage dioxygenase.
J.Inorg.Biochem., 83, 2001
|
|
7ZO1
 
 | | SpCas9 bound to CD34 off-target9 DNA substrate | | Descriptor: | CD34 off-target9 DNA non-target strand, CD34 off-target9 DNA target strand, CD34 sgRNA, ... | | Authors: | Pacesa, M, Jinek, M. | | Deposit date: | 2022-04-23 | | Release date: | 2022-10-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for Cas9 off-target activity.
Cell, 185, 2022
|
|
6I3U
 
 | |
5M8G
 
 | | Tubulin-MTD265 complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 5-(2-morpholin-4-yl-6-pyrrolidin-1-yl-pyrimidin-4-yl)-4-(trifluoromethyl)pyridin-2-amine, CALCIUM ION, ... | | Authors: | Bohnacker, T, Prota, A.E, Steinmetz, M.O, Wymann, M.P. | | Deposit date: | 2016-10-28 | | Release date: | 2017-02-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.147 Å) | | Cite: | Deconvolution of Buparlisib's mechanism of action defines specific PI3K and tubulin inhibitors for therapeutic intervention.
Nat Commun, 8, 2017
|
|
5MSL
 
 | |
5M7E
 
 | | Tubulin-BKM120 complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 5-[2,6-di(morpholin-4-yl)pyrimidin-4-yl]-4-(trifluoromethyl)pyridin-2-amine, CALCIUM ION, ... | | Authors: | Bohnacker, T, Prota, A.E, Steinmetz, M.O, Wymann, M.P. | | Deposit date: | 2016-10-27 | | Release date: | 2017-02-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.046 Å) | | Cite: | Deconvolution of Buparlisib's mechanism of action defines specific PI3K and tubulin inhibitors for therapeutic intervention.
Nat Commun, 8, 2017
|
|
5LXT
 
 | | Tubulin-Discodermolide complex | | Descriptor: | (+)-Discodermolide, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Prota, A.E, Steinmetz, M.O. | | Deposit date: | 2016-09-22 | | Release date: | 2017-03-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Basis of Microtubule Stabilization by Discodermolide.
Chembiochem, 18, 2017
|
|
