4YG7
 
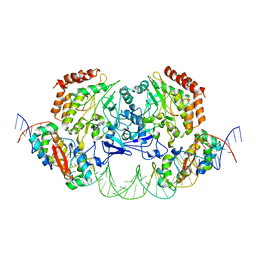 | | Structure of FL autorepression promoter complex | | Descriptor: | Antitoxin HipB, DNA (50-MER), Serine/threonine-protein kinase HipA | | Authors: | Schumacher, M.A. | | Deposit date: | 2015-02-25 | | Release date: | 2015-07-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.77 Å) | | Cite: | HipBA-promoter structures reveal the basis of heritable multidrug tolerance.
Nature, 524, 2015
|
|
4WZ6
 
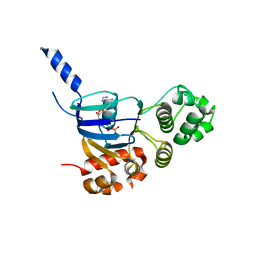 | | Human CFTR aa389-678 (NBD1), deltaF508 with three solubilizing mutations, bound ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cystic fibrosis transmembrane conductance regulator, MAGNESIUM ION | | Authors: | Byrnes, L.J, Hall, J. | | Deposit date: | 2014-11-18 | | Release date: | 2015-11-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Binding screen for cystic fibrosis transmembrane conductance regulator correctors finds new chemical matter and yields insights into cystic fibrosis therapeutic strategy.
Protein Sci., 25, 2016
|
|
1ACX
 
 | |
4IX8
 
 | |
3NKD
 
 | | Structure of CRISP-associated protein Cas1 from Escherichia coli str. K-12 | | Descriptor: | CRISPR-associated protein Cas1 | | Authors: | Nocek, B, Skarina, T, Beloglazova, N, Savchenko, A, Joachimiak, A, Yakunin, A. | | Deposit date: | 2010-06-18 | | Release date: | 2010-08-25 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A dual function of the CRISPR-Cas system in bacterial antivirus immunity and DNA repair.
Mol.Microbiol., 79, 2011
|
|
5B1W
 
 | |
4ZJ9
 
 | |
5AV7
 
 | | Crystal structure of Calsepa lectin in complex with bisected glycan | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)][2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)]methyl alpha-D-mannopyranoside, Lectin | | Authors: | Nagae, M, Yamaguchi, Y. | | Deposit date: | 2015-06-12 | | Release date: | 2016-04-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Atomic visualization of a flipped-back conformation of bisected glycans bound to specific lectins
Sci Rep, 6, 2016
|
|
7E6Q
 
 | | Crystal structure of influenza A virus neuraminidase N5 complexed with 4'-phenyl-1,2,3-triazolylated oseltamivir carboxylate | | Descriptor: | (3R,4R,5S)-4-acetamido-3-pentan-3-yloxy-5-(4-phenyl-1,2,3-triazol-1-yl)cyclohexene-1-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Wang, P.F, Babayemi, O.O, Li, C.N, Fu, L.F, Zhang, S.S, Qi, J.X, Lv, X, Li, X.B. | | Deposit date: | 2021-02-23 | | Release date: | 2021-03-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-based design of 5'-substituted 1,2,3-triazolylated oseltamivir derivatives as potent influenza neuraminidase inhibitors.
Rsc Adv, 11, 2021
|
|
3NKE
 
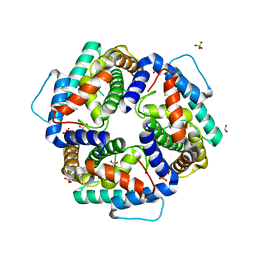 | | High resolution structure of the C-terminal domain CRISP-associated protein Cas1 from Escherichia coli str. K-12 | | Descriptor: | 1,2-ETHANEDIOL, SULFATE ION, SULFITE ION, ... | | Authors: | Nocek, B, Skarina, T, Beloglazova, N, Savchenko, A, Joachimiak, A, Yakunin, A. | | Deposit date: | 2010-06-18 | | Release date: | 2010-08-25 | | Last modified: | 2014-12-17 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | A dual function of the CRISPR-Cas system in bacterial antivirus immunity and DNA repair.
Mol.Microbiol., 79, 2011
|
|
5WCV
 
 | |
6HXW
 
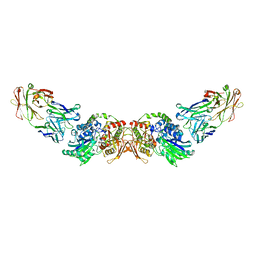 | | structure of human CD73 in complex with antibody IPH53 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 5'-nucleotidase, IPH53 heavy chain, ... | | Authors: | Roussel, A, Amigues, B. | | Deposit date: | 2018-10-18 | | Release date: | 2019-08-28 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Blocking Antibodies Targeting the CD39/CD73 Immunosuppressive Pathway Unleash Immune Responses in Combination Cancer Therapies.
Cell Rep, 27, 2019
|
|
5HI4
 
 | | Binding site elucidation and structure guided design of macrocyclic IL-17A antagonists | | Descriptor: | (9'S,17'R)-6'-chloro-N-methyl-9'-{[(1-methyl-1H-pyrazol-5-yl)carbonyl]amino}-10',19'-dioxo-2'-oxa-11',18'-diazaspiro[cyclopentane-1,21'-tetracyclo[20.2.2.2~12,15~.1~3,7~]nonacosane]-1'(24'),3'(29'),4',6',12',14',22',25',27'-nonaene-17'-carboxamide, CAT-2000 FAB heavy chain, CAT-2000 FAB light chain, ... | | Authors: | Liu, S. | | Deposit date: | 2016-01-11 | | Release date: | 2016-08-31 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Binding site elucidation and structure guided design of macrocyclic IL-17A antagonists.
Sci Rep, 6, 2016
|
|
5W1K
 
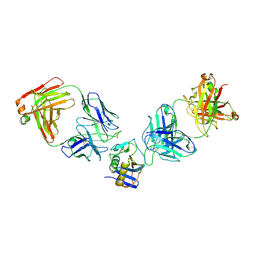 | | JUNV GP1 CR1-10 Fab CR1-28 Fab complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CR1-10 Fab heavy chain, ... | | Authors: | Raymond, D.D, Clark, L.E, Abraham, J. | | Deposit date: | 2017-06-03 | | Release date: | 2018-05-30 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.99 Å) | | Cite: | Vaccine-elicited receptor-binding site antibodies neutralize two New World hemorrhagic fever arenaviruses.
Nat Commun, 9, 2018
|
|
5VR9
 
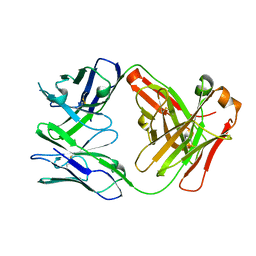 | | CH1/Ckappa Fab based on Matuzumab | | Descriptor: | CH1/Ckappa Fab heavy chain, CH1/Ckappa Fab light chain | | Authors: | Hendle, J. | | Deposit date: | 2017-05-10 | | Release date: | 2017-08-02 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Computational design of a specific heavy chain/ kappa light chain interface for expressing fully IgG bispecific antibodies.
Protein Sci., 26, 2017
|
|
5VSH
 
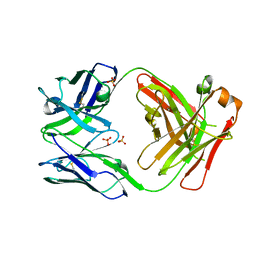 | | CH1/Clambda Fab based on Pertuzumab | | Descriptor: | CH1/Clambda Fab heavy chain, CH1/Clambda Fab light chain, SULFATE ION | | Authors: | Hendle, J. | | Deposit date: | 2017-05-11 | | Release date: | 2017-08-02 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Computational design of a specific heavy chain/ kappa light chain interface for expressing fully IgG bispecific antibodies.
Protein Sci., 26, 2017
|
|
5W1M
 
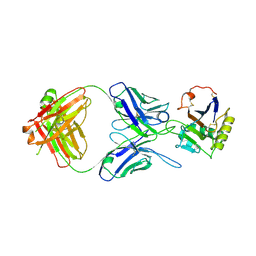 | | MACV GP1 CR1-07 Fab complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CR1-07 Fab heavy chain, ... | | Authors: | Raymond, D.D, Clark, L.E, Abraham, J. | | Deposit date: | 2017-06-03 | | Release date: | 2018-05-30 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.91 Å) | | Cite: | Vaccine-elicited receptor-binding site antibodies neutralize two New World hemorrhagic fever arenaviruses.
Nat Commun, 9, 2018
|
|
5B1X
 
 | |
5W1G
 
 | | CR1-07 unliganded Fab | | Descriptor: | CR1-07 Fab heavy chain, CR1-07 Fab light chain | | Authors: | Raymond, D.D, Clark, L.E, Abraham, J. | | Deposit date: | 2017-06-03 | | Release date: | 2018-05-30 | | Last modified: | 2020-01-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Vaccine-elicited receptor-binding site antibodies neutralize two New World hemorrhagic fever arenaviruses.
Nat Commun, 9, 2018
|
|
3N5C
 
 | | Crystal Structure of Arf6DELTA13 complexed with GDP | | Descriptor: | ADP-ribosylation factor 6, CHLORIDE ION, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Aizel, K, Biou, V, Cherfils, J. | | Deposit date: | 2010-05-25 | | Release date: | 2010-08-18 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | SAXS and X-ray crystallography suggest an unfolding model for the GDP/GTP conformational switch of the small GTPase Arf6.
J.Mol.Biol., 402, 2010
|
|
3N75
 
 | | X-ray Crystal Structure of the Escherichia coli Inducible Lysine Decarboxylase LdcI | | Descriptor: | GLYCEROL, GUANOSINE-5',3'-TETRAPHOSPHATE, HEXAETHYLENE GLYCOL, ... | | Authors: | Kanjee, U, Alexopoulos, E, Pai, E.F, Houry, W.A. | | Deposit date: | 2010-05-26 | | Release date: | 2011-02-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Linkage between the bacterial acid stress and stringent responses: the structure of the inducible lysine decarboxylase.
Embo J., 30, 2011
|
|
4L72
 
 | | Crystal structure of MERS-CoV complexed with human DPP4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Dipeptidyl peptidase 4, ... | | Authors: | Wang, X.Q, Wang, N.S. | | Deposit date: | 2013-06-13 | | Release date: | 2013-08-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.005 Å) | | Cite: | Structure of MERS-CoV spike receptor-binding domain complexed with human receptor DPP4
Cell Res., 23, 2013
|
|
1N4L
 
 | | A DNA analogue of the polypurine tract of HIV-1 | | Descriptor: | 5'-D(*CP*TP*TP*TP*TP*CP*TP*TP*TP*TP*AP*AP*AP*AP*AP*G)-3', 5'-D(*CP*TP*TP*TP*TP*TP*AP*AP*AP*AP*GP*AP*AP*AP*AP*G)-3', Reverse Transcriptase | | Authors: | Cote, M.L, Pflomm, M, Georgiadis, M.M. | | Deposit date: | 2002-10-31 | | Release date: | 2003-06-24 | | Last modified: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Staying Straight with A-tracts: A DNA Analog of the HIV-1 Polypurine Tract
J.Mol.Biol., 330, 2003
|
|
5C9I
 
 | |
1G0Z
 
 | |
