8E2M
 
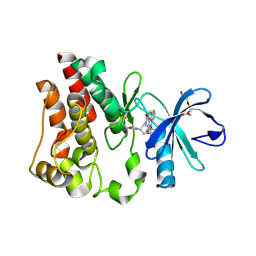 | | Bruton's tyrosine kinase (BTK) with compound 13 | | Descriptor: | (5P)-5-[4-methyl-6-(2-methylpropyl)pyridin-3-yl]-4-oxo-N-[(1R,2S)-2-propanamidocyclopentyl]-4,5-dihydro-3H-1-thia-3,5,8-triazaacenaphthylene-2-carboxamide, TRIETHYLENE GLYCOL, Tyrosine-protein kinase BTK | | Authors: | Alexander, R, Milligan, C.M. | | Deposit date: | 2022-08-15 | | Release date: | 2022-11-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.904 Å) | | Cite: | Discovery of JNJ-64264681: A Potent and Selective Covalent Inhibitor of Bruton's Tyrosine Kinase.
J.Med.Chem., 65, 2022
|
|
5NC8
 
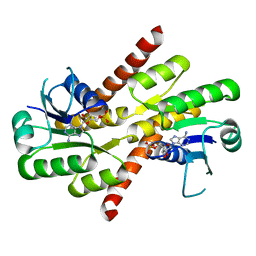 | | Shewanella denitrificans Kef CTD in AMP bound form | | Descriptor: | ADENOSINE MONOPHOSPHATE, Potassium efflux system protein | | Authors: | Pliotas, C, Naismith, J.H. | | Deposit date: | 2017-03-03 | | Release date: | 2017-04-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | Adenosine Monophosphate Binding Stabilizes the KTN Domain of the Shewanella denitrificans Kef Potassium Efflux System.
Biochemistry, 56, 2017
|
|
4LI4
 
 | |
4LM9
 
 | |
4LMC
 
 | |
4NL9
 
 | |
6G05
 
 | | RORGT (264-518;C455S) IN COMPLEX WITH INVERSE AGONIST "CPD-2" AND RIP140 PEPTIDE AT 1.90A | | Descriptor: | 2-(4-ethylsulfonylphenyl)-~{N}-[4-phenyl-5-(phenylcarbonyl)-1,3-thiazol-2-yl]ethanamide, Nuclear receptor ROR-gamma, Nuclear receptor-interacting protein 1 | | Authors: | Kallen, J. | | Deposit date: | 2018-03-16 | | Release date: | 2018-07-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Optimizing a Weakly Binding Fragment into a Potent ROR gamma t Inverse Agonist with Efficacy in an in Vivo Inflammation Model.
J. Med. Chem., 61, 2018
|
|
7V90
 
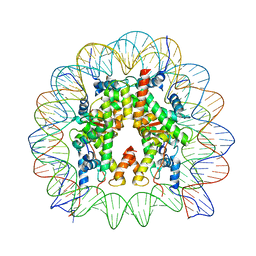 | | Telomeric mononucleosome | | Descriptor: | DNA (145-mer), Histone H2A type 1-B/E, Histone H2B type 1-K, ... | | Authors: | Soman, A. | | Deposit date: | 2021-08-24 | | Release date: | 2022-07-27 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Columnar structure of human telomeric chromatin.
Nature, 609, 2022
|
|
7VA4
 
 | | Telomeric tetranucleosome in open state | | Descriptor: | DNA (539-MER), Histone H2A type 1-B/E, Histone H2B type 1-K, ... | | Authors: | Soman, A. | | Deposit date: | 2021-08-27 | | Release date: | 2022-07-27 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (14 Å) | | Cite: | Columnar structure of human telomeric chromatin.
Nature, 609, 2022
|
|
7V9J
 
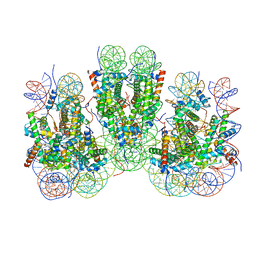 | | Telomeric trinucleosome | | Descriptor: | DNA (408-mer), Histone H2A type 1-B/E, Histone H2B type 1-K, ... | | Authors: | Soman, A. | | Deposit date: | 2021-08-25 | | Release date: | 2022-07-27 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (8 Å) | | Cite: | Columnar structure of human telomeric chromatin.
Nature, 609, 2022
|
|
7V9C
 
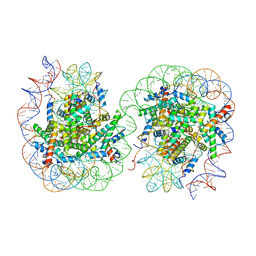 | | Telomeric Dinucleosome in open state | | Descriptor: | DNA (275-mer), Histone H2A type 1-B/E, Histone H2B type 1-K, ... | | Authors: | Soman, A. | | Deposit date: | 2021-08-24 | | Release date: | 2022-07-27 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Columnar structure of human telomeric chromatin.
Nature, 609, 2022
|
|
7V9S
 
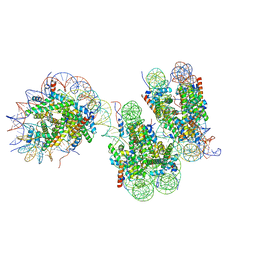 | | Telomeric trinucleosome in open state | | Descriptor: | DNA (408-MER), Histone H2A type 1-B/E, Histone H2B type 1-K, ... | | Authors: | Soman, A. | | Deposit date: | 2021-08-26 | | Release date: | 2022-07-27 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (11 Å) | | Cite: | Columnar structure of human telomeric chromatin.
Nature, 609, 2022
|
|
7V9K
 
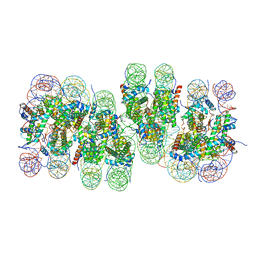 | | Telomeric tetranucleosome | | Descriptor: | DNA (539-mer), Histone H2A type 1-B/E, Histone H2B type 1-K, ... | | Authors: | Soman, A. | | Deposit date: | 2021-08-25 | | Release date: | 2022-07-27 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (8.1 Å) | | Cite: | Columnar structure of human telomeric chromatin.
Nature, 609, 2022
|
|
7V96
 
 | | Telomeric Dinucleosome | | Descriptor: | DNA (275-mer), Histone H2A type 1-B/E, Histone H2B type 1-K, ... | | Authors: | Soman, A. | | Deposit date: | 2021-08-24 | | Release date: | 2022-07-27 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.92 Å) | | Cite: | Columnar structure of human telomeric chromatin.
Nature, 609, 2022
|
|
8T85
 
 | | Structure of RssB bound to beryllofluoride | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, ... | | Authors: | Brugger, C, Schwartz, J, Deaconescu, A.M. | | Deposit date: | 2023-06-21 | | Release date: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Structure of phosphorylated-like RssB, the adaptor delivering sigma s to the ClpXP proteolytic machinery, reveals an interface switch for activation.
J.Biol.Chem., 299, 2023
|
|
4MI3
 
 | | Crystal structure of Gpb in complex with SUGAR (N-{(2R)-2-METHYL-3-[4-(PROPAN-2-YL)PHENYL]PROPANOYL}-BETA-D-GLUCOPYRANOSYLAMINE) (S21) | | Descriptor: | Glycogen phosphorylase, muscle form, N-{(2R)-2-methyl-3-[4-(propan-2-yl)phenyl]propanoyl}-beta-D-glucopyranosylamine | | Authors: | Kantsadi, L.A, Chatzileontiadou, S.M.D, Leonidas, D.D. | | Deposit date: | 2013-08-30 | | Release date: | 2014-07-23 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure based inhibitor design targeting glycogen phosphorylase b. Virtual screening, synthesis, biochemical and biological assessment of novel N-acyl-beta-d-glucopyranosylamines.
Bioorg.Med.Chem., 22, 2014
|
|
1KQY
 
 | | Hevamine Mutant D125A/E127A/Y183F in Complex with Penta-NAG | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Hevamine A, ... | | Authors: | Rozeboom, H.J, Dijkstra, B.W. | | Deposit date: | 2002-01-08 | | Release date: | 2002-01-23 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Expression and characterization of active site mutants of hevamine, a chitinase from the rubber tree Hevea brasiliensis.
Eur.J.Biochem., 269, 2002
|
|
1KR0
 
 | | Hevamine Mutant D125A/Y183F in Complex with Tetra-NAG | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hevamine A, SULFATE ION | | Authors: | Rozeboom, H.J, Dijkstra, B.W. | | Deposit date: | 2002-01-08 | | Release date: | 2002-01-23 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Expression and Characterization of Active Site Mutants of Hevamine, a
Chitinase from the Rubber Tree Hevea brasiliensis.
Eur.J.Biochem., 269, 2002
|
|
1KR1
 
 | | Hevamine Mutant D125A/E127A in Complex with Tetra-NAG | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hevamine A, SULFATE ION | | Authors: | Rozeboom, H.J, Dijkstra, B.W. | | Deposit date: | 2002-01-08 | | Release date: | 2002-01-23 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Expression and Characterization of Active Site Mutants of Hevamine, a
Chitinase from the Rubber Tree Hevea brasiliensis.
Eur.J.Biochem., 269, 2002
|
|
6WE5
 
 | |
8SV9
 
 | | Crystal structure of ULK1 kinase domain with inhibitor MR-2088 | | Descriptor: | (4P)-4-[(2P)-2-(1,2,5,6-tetrahydropyridin-3-yl)-1H-pyrrolo[2,3-b]pyridin-5-yl]-N-(2,2,2-trifluoroethyl)thiophene-2-carboxamide, 1,2-ETHANEDIOL, SULFATE ION, ... | | Authors: | Schonbrunn, E, Sun, L. | | Deposit date: | 2023-05-15 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Development of potent and selective ULK1/2 inhibitors based on 7-azaindole scaffold with favorable in vivo properties.
Eur.J.Med.Chem., 266, 2024
|
|
6WSA
 
 | |
6WSB
 
 | |
6WTS
 
 | | CryoEM structure of the C. sordellii lethal toxin TcsL in complex with SEMA6A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, SEMA6A, ... | | Authors: | Kucharska, I, Rubinstein, J.L, Julien, J.P. | | Deposit date: | 2020-05-03 | | Release date: | 2020-07-08 | | Last modified: | 2020-08-05 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Recognition of Semaphorin Proteins by P. sordellii Lethal Toxin Reveals Principles of Receptor Specificity in Clostridial Toxins.
Cell, 182, 2020
|
|
8T4D
 
 | | MD65 N332-GT5 SOSIP in complex with RM_N332_08 Fab and RM20A3 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, MD65 N332-GT5 SOSIP gp120, ... | | Authors: | Ozorowski, G, Torres, J.L, Ward, A.B. | | Deposit date: | 2023-06-09 | | Release date: | 2024-05-08 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Vaccine priming of rare HIV broadly neutralizing antibody precursors in nonhuman primates.
Science, 384, 2024
|
|
