6O4J
 
 | | Amyloid Beta KLVFFAENVGS 16-26 D23N Iowa mutation | | Descriptor: | Amyloid-beta precursor protein | | Authors: | Griner, S.L, Sawaya, M.R, Rodriguez, J.A, Cascio, D, Gonen, T. | | Deposit date: | 2019-02-28 | | Release date: | 2019-10-30 | | Last modified: | 2023-10-11 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.402 Å) | | Cite: | Structure based inhibitors of Amyloid Beta core suggest a common interface with Tau.
Elife, 8, 2019
|
|
5FOY
 
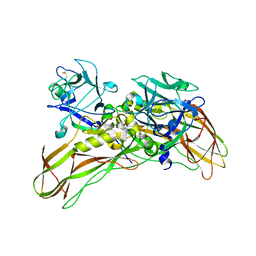 | | De novo structure of the binary mosquito larvicide BinAB at pH 7 | | Descriptor: | 41.9 KDA INSECTICIDAL TOXIN, LARVICIDAL TOXIN 51 KDA PROTEIN | | Authors: | Colletier, J.P, Sawaya, M.R, Gingery, M, Rodriguez, J.A, Cascio, D, Brewster, A.S, Michels-Clark, T, Boutet, S, Williams, G.J, Messerschmidt, M, DePonte, D.P, Sierra, R.G, Laksmono, H, Koglin, J.E, Hunter, M.S, W Park, H, Uervirojnangkoorn, M, Bideshi, D.L, Brunger, A.T, Federici, B.A, Sauter, N.K, Eisenberg, D.S. | | Deposit date: | 2015-11-26 | | Release date: | 2016-10-05 | | Last modified: | 2019-08-28 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | De Novo Phasing with X-Ray Laser Reveals Mosquito Larvicide Binab Structure.
Nature, 539, 2016
|
|
5G37
 
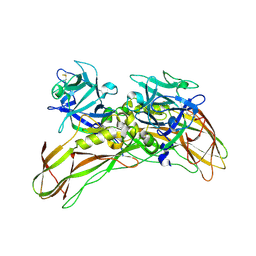 | | MR structure of the binary mosquito larvicide BinAB at pH 5 | | Descriptor: | 41.9 KDA INSECTICIDAL TOXIN, LARVICIDAL TOXIN 51 KDA PROTEIN | | Authors: | Colletier, J.P, Sawaya, M.R, Gingery, M, Rodriguez, J.A, Cascio, D, Brewster, A.S, Michels-Clark, T, Boutet, S, Williams, G.J, Messerschmidt, M, DePonte, D.P, Sierra, R.G, Laksmono, H, Koglin, J.E, Hunter, M.S, W Park, H, Uervirojnangkoorn, M, Bideshi, D.L, Brunger, A.T, Federici, B.A, Sauter, N.K, Eisenberg, D.S. | | Deposit date: | 2016-04-24 | | Release date: | 2016-10-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | De Novo Phasing with X-Ray Laser Reveals Mosquito Larvicide Binab Structure.
Nature, 539, 2016
|
|
2Z9H
 
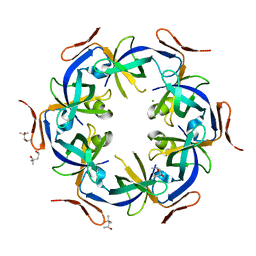 | | Ethanolamine utilization protein, EutN | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, CHLORIDE ION, Ethanolamine utilization protein eutN | | Authors: | Tanaka, S, Sawaya, M.R, Yeates, T.O. | | Deposit date: | 2007-09-20 | | Release date: | 2007-10-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | The crystal structure of ethanolamine utilization protein EutN from E. coli
To be Published
|
|
2Y5Y
 
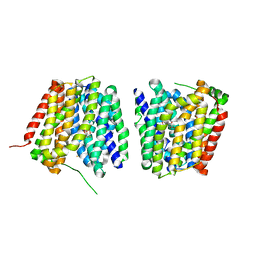 | | Crystal structure of LacY in complex with an affinity inactivator | | Descriptor: | 2-sulfanylethyl beta-D-galactopyranoside, BARIUM ION, LACTOSE PERMEASE | | Authors: | Chaptal, V, Kwon, S, Sawaya, M.R, Guan, L, Kaback, H.R, Abramson, J. | | Deposit date: | 2011-01-19 | | Release date: | 2011-06-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.38 Å) | | Cite: | Crystal Structure of Lactose Permease in Complex with an Affinity Inactivator Yields Unique Insight Into Sugar Recognition.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
2Y2A
 
 | | Structure of segment KLVFFA from the amyloid-beta peptide (Ab, residues 16-21), alternate polymorph I | | Descriptor: | ACETATE ION, AMYLOID BETA A4 PROTEIN | | Authors: | Colletier, J, Laganowsky, A, Sawaya, M.R, Eisenberg, D. | | Deposit date: | 2010-12-14 | | Release date: | 2011-10-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Molecular Basis for Amyloid-{Beta} Polymorphism.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
2Y3L
 
 | | Structure of segment MVGGVVIA from the amyloid-beta peptide (Ab, residues 35-42), alternate polymorph 2 | | Descriptor: | AMYLOID BETA A4 PROTEIN | | Authors: | Colletier, J.P, Laganowsky, A, Sawaya, M.R, Eisenberg, D. | | Deposit date: | 2010-12-21 | | Release date: | 2011-11-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Molecular Basis for Amyloid-{Beta} Polymorphism.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
2Y29
 
 | | Structure of segment KLVFFA from the amyloid-beta peptide (Ab, residues 16-21), alternate polymorph III | | Descriptor: | AMYLOID BETA A4 PROTEIN | | Authors: | Colletier, J, Laganowsky, A, Sawaya, M.R, Eisenberg, D. | | Deposit date: | 2010-12-14 | | Release date: | 2011-10-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular Basis for Amyloid-{Beta} Polymorphism.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
2Y3K
 
 | | Structure of segment MVGGVVIA from the amyloid-beta peptide (Ab, residues 35-42), alternate polymorph 1 | | Descriptor: | AMYLOID BETA A4 PROTEIN | | Authors: | Colletier, J.P, Laganowsky, A, Sawaya, M.R, Eisenberg, D. | | Deposit date: | 2010-12-21 | | Release date: | 2011-11-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular Basis for Amyloid-{Beta} Polymorphism.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
2Y3J
 
 | | Structure of segment AIIGLM from the amyloid-beta peptide (Ab, residues 30-35) | | Descriptor: | AMYLOID BETA A4 PROTEIN | | Authors: | Colletier, J.P, Laganowsky, A, Sawaya, M.R, Eisenberg, D. | | Deposit date: | 2010-12-21 | | Release date: | 2011-11-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Molecular Basis for Amyloid-{Beta} Polymorphism.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3BN4
 
 | | Carboxysome Subunit, CcmK1 | | Descriptor: | Carbon dioxide-concentrating mechanism protein ccmK homolog 1, SULFATE ION | | Authors: | Tanaka, S, Sawaya, M.R, Yeates, T.O. | | Deposit date: | 2007-12-13 | | Release date: | 2008-03-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Atomic-level models of the bacterial carboxysome shell.
Science, 319, 2008
|
|
3BQ7
 
 | |
8T0B
 
 | |
8T1N
 
 | | Micro-ED Structure of a Novel Domain of Unknown Function Solved with AlphaFold | | Descriptor: | DUF1842 domain-containing protein | | Authors: | Miller, J.E, Cascio, D, Sawaya, M.R, Cannon, K.A, Rodriguez, J.A, Yeates, T.O. | | Deposit date: | 2023-06-02 | | Release date: | 2024-01-17 | | Last modified: | 2024-04-10 | | Method: | ELECTRON CRYSTALLOGRAPHY (3 Å) | | Cite: | AlphaFold-assisted structure determination of a bacterial protein of unknown function using X-ray and electron crystallography.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
8T1M
 
 | | Novel Domain of Unknown Function Solved with AlphaFold | | Descriptor: | DUF1842 domain-containing protein | | Authors: | Miller, J.E, Agdanowski, M.P, Cascio, D, Sawaya, M.R, Yeates, T.O. | | Deposit date: | 2023-06-02 | | Release date: | 2024-01-17 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | AlphaFold-assisted structure determination of a bacterial protein of unknown function using X-ray and electron crystallography.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
8SPA
 
 | | Structural insights into cellular control of the human CPEB3 prion, functionally regulated by a labile-amyloid-forming segment | | Descriptor: | Cytoplasmic polyadenylation element-binding protein 3 | | Authors: | Flores, M.D, Sawaya, M.R, Boyer, D.R, Zink, S, Fioriti, L, Rodriguez, J.A. | | Deposit date: | 2023-05-02 | | Release date: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure of a reversible amyloid fibril formed by the CPEB3 prion-like domain reveals a core sequence involved in translational regulation
To Be Published
|
|
8G47
 
 | | KRAS G12C complex with GDP and AMG 510 imaged on a cryo-EM imaging scaffold | | Descriptor: | AMG 510 (bound form), GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Castells-Graells, R, Sawaya, M.R, Yeates, T.O. | | Deposit date: | 2023-02-08 | | Release date: | 2023-08-09 | | Last modified: | 2023-09-27 | | Method: | ELECTRON MICROSCOPY (3.19 Å) | | Cite: | Cryo-EM structure determination of small therapeutic protein targets at 3 angstrom -resolution using a rigid imaging scaffold.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8G3K
 
 | |
8G4H
 
 | | KRAS G13C complex with GDP imaged on a cryo-EM imaging scaffold | | Descriptor: | GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Castells-Graells, R, Sawaya, M.R, Yeates, T.O. | | Deposit date: | 2023-02-09 | | Release date: | 2023-08-09 | | Last modified: | 2023-09-27 | | Method: | ELECTRON MICROSCOPY (2.87 Å) | | Cite: | Cryo-EM structure determination of small therapeutic protein targets at 3 angstrom -resolution using a rigid imaging scaffold.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8G4E
 
 | |
8G42
 
 | | KRAS G12C complex with GDP imaged on a cryo-EM imaging scaffold | | Descriptor: | GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Castells-Graells, R, Sawaya, M.R, Yeates, T.O. | | Deposit date: | 2023-02-08 | | Release date: | 2023-08-09 | | Last modified: | 2023-09-27 | | Method: | ELECTRON MICROSCOPY (3.02 Å) | | Cite: | Cryo-EM structure determination of small therapeutic protein targets at 3 angstrom -resolution using a rigid imaging scaffold.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8G4F
 
 | | KRAS G12V complex with GDP imaged on a cryo-EM imaging scaffold | | Descriptor: | GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Castells-Graells, R, Sawaya, M.R, Yeates, T.O. | | Deposit date: | 2023-02-09 | | Release date: | 2023-08-09 | | Last modified: | 2023-09-27 | | Method: | ELECTRON MICROSCOPY (2.91 Å) | | Cite: | Cryo-EM structure determination of small therapeutic protein targets at 3 angstrom -resolution using a rigid imaging scaffold.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8G2V
 
 | | Cryo-EM structure of recombinant human LECT2 amyloid fibril core | | Descriptor: | Leukocyte cell-derived chemotaxin-2 | | Authors: | Richards, L.S, Flores, M.D, Zink, S, Schibrowsky, N.A, Sawaya, M.R, Rodriguez, J.A. | | Deposit date: | 2023-02-06 | | Release date: | 2023-08-16 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.715 Å) | | Cite: | Cryo-EM structure of a human LECT2 amyloid fibril reveals a network of polar ladders at its core.
Structure, 31, 2023
|
|
6E6Z
 
 | | Structure of Wild Type Human Transthyretin in Complex with Tafamidis | | Descriptor: | 2-(3,5-dichlorophenyl)-1,3-benzoxazole-6-carboxylic acid, Transthyretin | | Authors: | Chung, K, Saelices, L, Sawaya, M.R, Cascio, D, Eisenberg, D. | | Deposit date: | 2018-07-25 | | Release date: | 2019-07-31 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural Variants of Transthyretin
To Be Published
|
|
6E77
 
 | | Structure of Human Transthyretin Asp38Ala Mutant in Complex with Tafamidis | | Descriptor: | 2-(3,5-dichlorophenyl)-1,3-benzoxazole-6-carboxylic acid, Transthyretin | | Authors: | Saelices, L, Chung, K, Sawaya, M.R, Cascio, D, Eisenberg, D. | | Deposit date: | 2018-07-25 | | Release date: | 2019-07-31 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural Variants of Transthyretin
To Be Published
|
|
