2B1G
 
 | | Crystal structures of transition state analogue inhibitors of inosine monophosphate cyclohydrolase | | Descriptor: | 7-(3,4-DIHYDROXY-5R-HYDROXYMETHYLTETRAHYDROFURAN-2-YL)-2,2-DIOXO-1,2R,3R,7-TETRAHYDRO-2L6-IMIDAZO[4,5-C][1,2,6]THIADIAZIN-4S-ONE, Bifunctional purine biosynthesis protein PURH, PHOSPHATE ION, ... | | Authors: | Xu, L, Chong, Y, Hwang, I, D'Onofrio, A, Amore, K, Beardsley, G.P, Li, C, Olson, A.J, Boger, D.L, Wilson, I.A. | | Deposit date: | 2005-09-15 | | Release date: | 2006-11-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-based Design, Synthesis, Evaluation, and Crystal Structures of Transition State Analogue Inhibitors of Inosine Monophosphate Cyclohydrolase.
J.Biol.Chem., 282, 2007
|
|
2B1I
 
 | | crystal structures of transition state analogue inhibitors of inosine monophosphate cyclohydrolase | | Descriptor: | Bifunctional purine biosynthesis protein PURH, POTASSIUM ION, [3,4-DIHYDROXY-5R-(2,2,4-TRIOXO-1,2R,3S,4R-TETRAHYDRO-2L6-IMIDAZO[4,5-C][1,2,6]THIADIAZIN-7-YL)TETRAHYDROFURAN-2-YL]METHYL DIHYDROGEN PHOSPHATE | | Authors: | Xu, L, Chong, Y, Hwang, I, D'Onofrio, A, Amore, K, Beardsley, G.P, Li, C, Olson, A.J, Boger, D.L, Wilson, I.A. | | Deposit date: | 2005-09-15 | | Release date: | 2006-11-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Structure-based Design, Synthesis, Evaluation, and Crystal Structures of Transition State Analogue Inhibitors of Inosine Monophosphate Cyclohydrolase.
J.Biol.Chem., 282, 2007
|
|
5B88
 
 | | RRM-like domain of DEAD-box protein, CsdA | | Descriptor: | ATP-dependent RNA helicase DeaD | | Authors: | Xu, L, Peng, J, Zhang, J, Wu, J, Tang, Y, Shi, Y. | | Deposit date: | 2016-06-13 | | Release date: | 2017-05-31 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Insights into the Structure of Dimeric RNA Helicase CsdA and Indispensable Role of Its C-Terminal Regions.
Structure, 25, 2017
|
|
1N7V
 
 | | THE RECEPTOR-BINDING PROTEIN P2 OF BACTERIOPHAGE PRD1: CRYSTAL FORM III | | Descriptor: | ACETATE ION, Adsorption protein P2, CALCIUM ION | | Authors: | Xu, L, Benson, S.D, Butcher, S.J, Bamford, D.H, Burnett, R.M. | | Deposit date: | 2002-11-18 | | Release date: | 2003-04-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Receptor Binding Protein P2 of PRD1, a
Virus Targeting Antibiotic-Resistant Bacteria,
Has a Novel Fold Suggesting Multiple Functions.
Structure, 11, 2003
|
|
1N7U
 
 | | THE RECEPTOR-BINDING PROTEIN P2 OF BACTERIOPHAGE PRD1: CRYSTAL FORM I | | Descriptor: | ACETATE ION, Adsorption protein P2, CALCIUM ION | | Authors: | Xu, L, Benson, S.D, Butcher, S.J, Bamford, D.H, Burnett, R.M. | | Deposit date: | 2002-11-18 | | Release date: | 2003-04-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The Receptor Binding Protein P2 of PRD1, a
Virus Targeting Antibiotic-Resistant Bacteria,
Has a Novel Fold Suggesting Multiple Functions.
Structure, 11, 2003
|
|
8IRL
 
 | | Apo state of Arabidopsis AZG1 at pH 7.4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Adenine/guanine permease AZG1 | | Authors: | Xu, L, Guo, J. | | Deposit date: | 2023-03-19 | | Release date: | 2024-01-17 | | Last modified: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structures and mechanisms of the Arabidopsis cytokinin transporter AZG1.
Nat.Plants, 10, 2024
|
|
8IRN
 
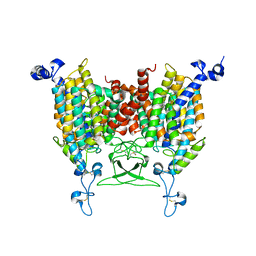 | | 6-BAP bound state of Arabidopsis AZG1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Adenine/guanine permease AZG1, N-BENZYL-9H-PURIN-6-AMINE | | Authors: | Xu, L, Guo, J. | | Deposit date: | 2023-03-19 | | Release date: | 2024-01-17 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structures and mechanisms of the Arabidopsis cytokinin transporter AZG1.
Nat.Plants, 10, 2024
|
|
8IRP
 
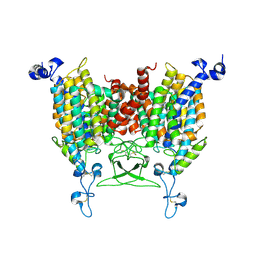 | | kinetin bound state of Arabidopsis AZG1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Adenine/guanine permease AZG1, N-(FURAN-2-YLMETHYL)-7H-PURIN-6-AMINE | | Authors: | Xu, L, Guo, J. | | Deposit date: | 2023-03-19 | | Release date: | 2024-01-17 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structures and mechanisms of the Arabidopsis cytokinin transporter AZG1.
Nat.Plants, 10, 2024
|
|
8IRM
 
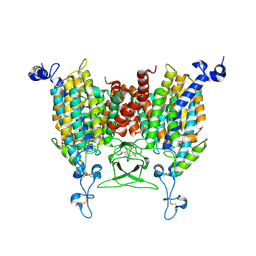 | | Endogenous substrate adenine bound state of Arabidopsis AZG1 at pH 5.5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ADENINE, Adenine/guanine permease AZG1 | | Authors: | Xu, L, Guo, J. | | Deposit date: | 2023-03-19 | | Release date: | 2024-01-17 | | Last modified: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structures and mechanisms of the Arabidopsis cytokinin transporter AZG1.
Nat.Plants, 10, 2024
|
|
8IRO
 
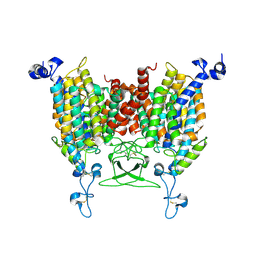 | | trans-Zeatin bound state of Arabidopsis AZG1 at pH7.4 | | Descriptor: | (2E)-2-methyl-4-(9H-purin-6-ylamino)but-2-en-1-ol, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Adenine/guanine permease AZG1 | | Authors: | Xu, L, Guo, J. | | Deposit date: | 2023-03-19 | | Release date: | 2024-01-17 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structures and mechanisms of the Arabidopsis cytokinin transporter AZG1.
Nat.Plants, 10, 2024
|
|
5GI4
 
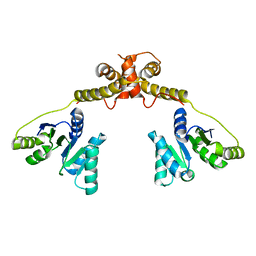 | | DEAD-box RNA helicase | | Descriptor: | ATP-dependent RNA helicase DeaD | | Authors: | Xu, L, Wang, L, Li, F, Wu, L, Shi, Y. | | Deposit date: | 2016-06-22 | | Release date: | 2017-05-31 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.244 Å) | | Cite: | Insights into the Structure of Dimeric RNA Helicase CsdA and Indispensable Role of Its C-Terminal Regions.
Structure, 25, 2017
|
|
3HVN
 
 | | Crystal structure of cytotoxin protein suilysin from Streptococcus suis | | Descriptor: | 1,1,1,3,3,3-hexafluoropropan-2-ol, HEPTANE-1,2,3-TRIOL, Hemolysin | | Authors: | Xu, L, Huang, B, Du, H, Zhang, C.X, Xu, J, Li, X, Rao, Z. | | Deposit date: | 2009-06-16 | | Release date: | 2010-03-02 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.852 Å) | | Cite: | Crystal structure of cytotoxin protein suilysin from Streptococcus suis.
Protein Cell, 1, 2010
|
|
5GJU
 
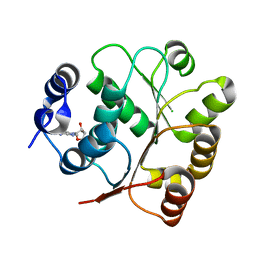 | | DEAD-box RNA helicase | | Descriptor: | ADENOSINE MONOPHOSPHATE, ATP-dependent RNA helicase DeaD | | Authors: | Xu, L, Li, F, Wang, L, Shi, Y. | | Deposit date: | 2016-07-02 | | Release date: | 2017-05-31 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Insights into the Structure of Dimeric RNA Helicase CsdA and Indispensable Role of Its C-Terminal Regions.
Structure, 25, 2017
|
|
2IU3
 
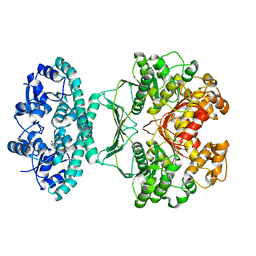 | | Crystal structures of transition state analogue inhibitors of inosine monophosphate cyclohydrolase | | Descriptor: | 1,5-DIHYDROIMIDAZO[4,5-C][1,2,6]THIADIAZIN-4(3H)-ONE 2,2-DIOXIDE, BIFUNCTIONAL PURINE BIOSYNTHESIS PROTEIN PURH, POTASSIUM ION | | Authors: | Xu, L, Chong, Y, Hwang, I, D'Onofrio, A, Amore, K, Beardsley, G.P, Li, C, Olson, A.J, Boger, D.L, Wilson, I.A. | | Deposit date: | 2006-05-27 | | Release date: | 2007-02-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure-based design, synthesis, evaluation, and crystal structures of transition state analogue inhibitors of inosine monophosphate cyclohydrolase.
J. Biol. Chem., 282, 2007
|
|
2IU0
 
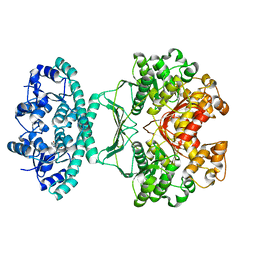 | | crystal structures of transition state analogue inhibitors of inosine monophosphate cyclohydrolase | | Descriptor: | 1,5-DIHYDROIMIDAZO[4,5-C][1,2,6]THIADIAZIN-4(3H)-ONE 2,2-DIOXIDE, BIFUNCTIONAL PURINE BIOSYNTHESIS PROTEIN PURH, POTASSIUM ION | | Authors: | Xu, L, Chong, Y, Hwang, I, Onofrio, A.D, Amore, K, Beardsley, G.P, Li, C, Olson, A.J, Boger, D.L, Wilson, I.A. | | Deposit date: | 2006-05-26 | | Release date: | 2007-02-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Structure-Based Design, Synthesis, Evaluation, and Crystal Structures of Transition State Analogue Inhibitors of Inosine Monophosphate Cyclohydrolase
J.Biol.Chem., 282, 2007
|
|
8T2T
 
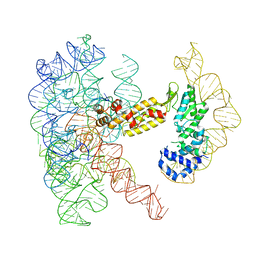 | | Structure of a group II intron ribonucleoprotein in the post-ligation (post-2F) state | | Descriptor: | AMMONIUM ION, Group II intron reverse transcriptase/maturase, MAGNESIUM ION, ... | | Authors: | Xu, L, Liu, T, Chung, K, Pyle, A.M. | | Deposit date: | 2023-06-06 | | Release date: | 2023-11-22 | | Last modified: | 2024-01-03 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insights into intron catalysis and dynamics during splicing.
Nature, 624, 2023
|
|
8T2R
 
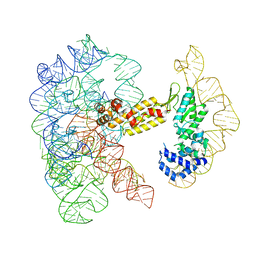 | | Structure of a group II intron ribonucleoprotein in the pre-ligation (pre-2F) state | | Descriptor: | 5'exon, AMMONIUM ION, CALCIUM ION, ... | | Authors: | Xu, L, Liu, T, Chung, K, Pyle, A.M. | | Deposit date: | 2023-06-06 | | Release date: | 2023-11-22 | | Last modified: | 2024-01-03 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insights into intron catalysis and dynamics during splicing.
Nature, 624, 2023
|
|
8T2S
 
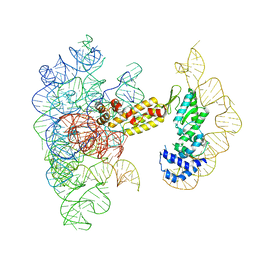 | | Structure of a group II intron ribonucleoprotein in the pre-branching (pre-1F) state | | Descriptor: | AMMONIUM ION, CALCIUM ION, Group II intron reverse transcriptase/maturase, ... | | Authors: | Xu, L, Liu, T, Chung, K, Pyle, A.M. | | Deposit date: | 2023-06-06 | | Release date: | 2023-11-22 | | Last modified: | 2024-01-03 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insights into intron catalysis and dynamics during splicing.
Nature, 624, 2023
|
|
1THZ
 
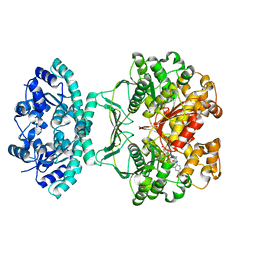 | | Crystal Structure of Avian AICAR Transformylase in Complex with a Novel Inhibitor Identified by Virtual Ligand Screening | | Descriptor: | 2-{(E)-[5-HYDROXY-3-METHYL-1-(2-METHYL-4-SULFOPHENYL)-1H-PYRAZOL-4-YL]DIAZENYL}-4-SULFOBENZOIC ACID, Bifunctional purine biosynthesis protein PURH, POTASSIUM ION | | Authors: | Xu, L, Li, C, Olson, A.J, Wilson, I.A. | | Deposit date: | 2004-06-01 | | Release date: | 2004-09-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of avian aminoimidazole-4-carboxamide ribonucleotide transformylase in complex with a novel non-folate inhibitor identified by virtual ligand screening.
J.Biol.Chem., 279, 2004
|
|
2INY
 
 | |
3T42
 
 | | Human aldose reductase in complex with a nitrile-containing IDD inhibitor | | Descriptor: | Aldose reductase, CITRIC ACID, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Xu, L, Cohen, A.E, Boxer, S.G. | | Deposit date: | 2011-07-25 | | Release date: | 2011-10-05 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Electrostatic Fields near the Active Site of Human Aldose Reductase: 2. New Inhibitors and Complications Caused by Hydrogen Bonds.
Biochemistry, 50, 2011
|
|
5GH9
 
 | | Crystal structure of CBP Bromodomain with H3K56ac peptide | | Descriptor: | CREB-binding protein, Histone H3 | | Authors: | Xu, L. | | Deposit date: | 2016-06-19 | | Release date: | 2017-06-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.451 Å) | | Cite: | Structural insight into CBP bromodomain-mediated recognition of acetylated histone H3K56ac
FEBS J., 2017
|
|
3FYM
 
 | | The 1A structure of YmfM, a putative DNA-binding membrane protein from Staphylococcus aureus | | Descriptor: | Putative uncharacterized protein, ZINC ION | | Authors: | Xu, L, Sedelnikova, S.E, Baker, P.J, Rice, D.W. | | Deposit date: | 2009-01-22 | | Release date: | 2010-02-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | The 1A structure of YmfM, a putative DNA-binding membrane protein from Staphylococcus aureus
To be Published
|
|
2ODM
 
 | | Crystal structure of S. aureus YlaN, an essential leucine rich protein involved in the control of cell shape | | Descriptor: | UPF0358 protein MW0995 | | Authors: | Xu, L, Sedelnikova, S.E, Baker, P.J, Errington, J, Hunt, A, Rice, D.W. | | Deposit date: | 2006-12-23 | | Release date: | 2007-06-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Crystal structure of S. aureus YlaN, an essential leucine rich protein involved in the control of cell shape.
Proteins, 68, 2007
|
|
8WO7
 
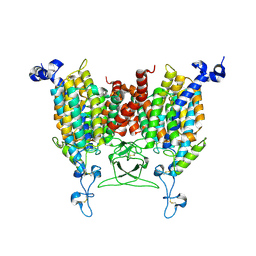 | | Apo state of Arabidopsis AZG1 T440Y | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Adenine/guanine permease AZG1 | | Authors: | Xu, L, Guo, J. | | Deposit date: | 2023-10-06 | | Release date: | 2024-01-10 | | Last modified: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structures and mechanisms of the Arabidopsis cytokinin transporter AZG1.
Nat.Plants, 10, 2024
|
|
