1W92
 
 | | The structure of carbomonoxy murine neuroglobin reveals a heme- sliding mechanism for affinity regulation | | Descriptor: | CARBON MONOXIDE, NEUROGLOBIN, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Vallone, B, Nienhaus, K, Matthes, A, Brunori, M, Nienhaus, G.U. | | Deposit date: | 2004-10-05 | | Release date: | 2004-11-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Structure of Carbonmonoxy Neuroglobin Reveals a Heme-Sliding Mechanism for Control of Ligand Affinity
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1Q1F
 
 | | Crystal structure of murine neuroglobin | | Descriptor: | Neuroglobin, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Vallone, B, Nienhaus, K, Matthes, K, Brunori, M, Nienhaus, G.U. | | Deposit date: | 2003-07-19 | | Release date: | 2004-06-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The structure of murine neuroglobin: Novel pathways for ligand migration and binding.
Proteins, 56, 2004
|
|
1GLI
 
 | |
4MU5
 
 | | Crystal structure of murine neuroglobin mutant M144W | | Descriptor: | Neuroglobin, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Vallone, B, Avella, G, Savino, C, Ardiccioni, C, Brunori, M. | | Deposit date: | 2013-09-20 | | Release date: | 2014-06-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Engineering the internal cavity of neuroglobin demonstrates the role of the haem-sliding mechanism.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3ZKP
 
 | |
1QI8
 
 | | DEOXYGENATED STRUCTURE OF A DISTAL POCKET HEMOGLOBIN MUTANT | | Descriptor: | HEMOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Miele, A.E, Vallone, B, Santanche, S, Travaglini-Allocatelli, C, Bellelli, A, Brunori, M. | | Deposit date: | 1999-06-07 | | Release date: | 1999-07-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Modulation of ligand binding in engineered human hemoglobin distal pocket.
J.Mol.Biol., 290, 1999
|
|
4XE3
 
 | | OleP, the cytochrome P450 epoxidase from Streptomyces antibioticus involved in Oleandomycin biosynthesis: functional analysis and crystallographic structure in complex with clotrimazole. | | Descriptor: | 1-[(2-CHLOROPHENYL)(DIPHENYL)METHYL]-1H-IMIDAZOLE, Cytochrome P-450, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Montemiglio, L.C, Parisi, G, Scaglione, A, Savino, C, Vallone, B. | | Deposit date: | 2014-12-22 | | Release date: | 2015-11-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Functional analysis and crystallographic structure of clotrimazole bound OleP, a cytochrome P450 epoxidase from Streptomyces antibioticus involved in oleandomycin biosynthesis.
Biochim.Biophys.Acta, 1860, 2015
|
|
7OHD
 
 | | CRYSTAL STRUCTURE OF FERRIC MURINE NEUROGLOBIN CDLESS MUTANT | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, ISOPROPYL ALCOHOL, ... | | Authors: | Exertier, C, Freda, I, Montemiglio, L.C, Savino, C, Cerutti, G, Gugole, E, Vallone, B. | | Deposit date: | 2021-05-10 | | Release date: | 2022-07-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Probing the Role of Murine Neuroglobin CDloop-D-Helix Unit in CO Ligand Binding and Structural Dynamics.
Acs Chem.Biol., 17, 2022
|
|
2XFH
 
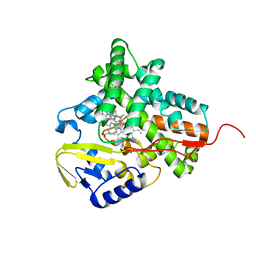 | | Structure of cytochrome P450 EryK cocrystallized with inhibitor clotrimazole. | | Descriptor: | 1-[(2-CHLOROPHENYL)(DIPHENYL)METHYL]-1H-IMIDAZOLE, DIMETHYL SULFOXIDE, ERYTHROMYCIN B/D C-12 HYDROXYLASE, ... | | Authors: | Savino, C, Montemiglio, L.C, Gianni, S, Vallone, B. | | Deposit date: | 2010-05-24 | | Release date: | 2010-09-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Azole Drugs Trap Cytochrome P450 Eryk in Alternative Conformational States.
Biochemistry, 49, 2010
|
|
6ZHZ
 
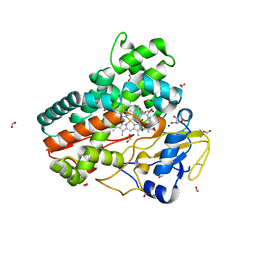 | | OleP-oleandolide(DEO) in high salt crystallization conditions | | Descriptor: | (3~{R},4~{S},5~{R},6~{S},7~{S},9~{S},11~{R},12~{S},13~{R},14~{R})-3,5,7,9,11,13,14-heptamethyl-4,6,12-tris(oxidanyl)-1-oxacyclotetradecane-2,10-dione, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Cytochrome P-450, ... | | Authors: | Montemiglio, L.C, Savino, C, Vallone, B, Parisi, G, Cecchetti, C. | | Deposit date: | 2020-06-24 | | Release date: | 2020-10-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Dissecting the Cytochrome P450 OleP Substrate Specificity: Evidence for a Preferential Substrate.
Biomolecules, 10, 2020
|
|
6ZI2
 
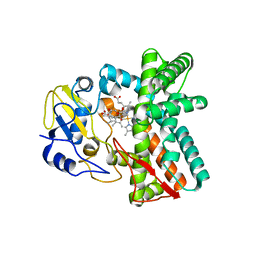 | | OleP-oleandolide(DEO) in low salt crystallization conditions | | Descriptor: | (3~{R},4~{S},5~{R},6~{S},7~{S},9~{S},11~{R},12~{S},13~{R},14~{R})-3,5,7,9,11,13,14-heptamethyl-4,6,12-tris(oxidanyl)-1-oxacyclotetradecane-2,10-dione, Cytochrome P-450, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Savino, C, Montemiglio, L.C, Vallone, B, Parisi, G, Cecchetti, C. | | Deposit date: | 2020-06-24 | | Release date: | 2020-10-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.93 Å) | | Cite: | Dissecting the Cytochrome P450 OleP Substrate Specificity: Evidence for a Preferential Substrate.
Biomolecules, 10, 2020
|
|
1F63
 
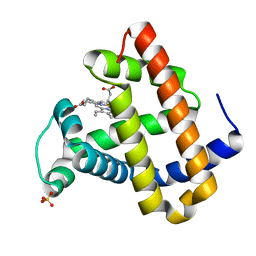 | | CRYSTAL STRUCTURE OF DEOXY SPERM WHALE MYOGLOBIN MUTANT Y(B10)Q(E7)R(E10) | | Descriptor: | MYOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Brunori, M, Cutruzzola, F, Savino, C, Travaglini-Allocatelli, C, Vallone, B, Gibson, Q.H. | | Deposit date: | 2000-06-20 | | Release date: | 2000-07-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural dynamics of ligand diffusion in the protein matrix: A study on a new myoglobin mutant Y(B10) Q(E7) R(E10).
Biophys.J., 76, 1999
|
|
1F65
 
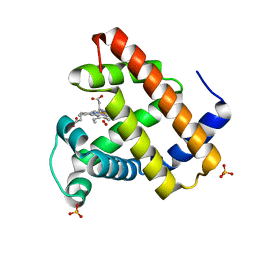 | | CRYSTAL STRUCTURE OF OXY SPERM WHALE MYOGLOBIN MUTANT Y(B10)Q(E7)R(E10) | | Descriptor: | MYOGLOBIN, OXYGEN MOLECULE, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Brunori, M, Cutruzzola, F, Savino, C, Travaglini-Allocatelli, C, Vallone, B, Gibson, Q.H. | | Deposit date: | 2000-06-20 | | Release date: | 2000-07-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural dynamics of ligand diffusion in the protein matrix: A study on a new myoglobin mutant Y(B10) Q(E7) R(E10).
Biophys.J., 76, 1999
|
|
1J7Y
 
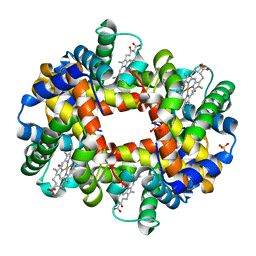 | | Crystal structure of partially ligated mutant of HbA | | Descriptor: | CARBON MONOXIDE, Hemoglobin, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Miele, A.E, Draghi, F, Arcovito, A, Bellelli, A, Brunori, M, Travaglini-Allocatelli, C, Vallone, B. | | Deposit date: | 2001-05-19 | | Release date: | 2002-02-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Control of heme reactivity by diffusion: structural basis and functional characterization in hemoglobin mutants.
Biochemistry, 40, 2001
|
|
1LQ9
 
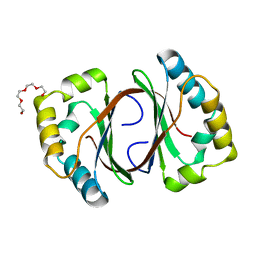 | | Crystal Structure of a Monooxygenase from the Gene ActVA-Orf6 of Streptomyces coelicolor Strain A3(2) | | Descriptor: | ACTVA-ORF6 MONOOXYGENASE, TETRAETHYLENE GLYCOL | | Authors: | Sciara, G, Kendrew, S.G, Miele, A.E, Marsh, N.G, Federici, L, Malatesta, F, Schimperna, G, Savino, C, Vallone, B. | | Deposit date: | 2002-05-09 | | Release date: | 2003-01-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | The structure of ActVA-Orf6, a novel type of monooxygenase involved in actinorhodin biosynthesis
EMBO J., 22, 2003
|
|
6I40
 
 | | Crystal structure of murine neuroglobin bound to CO at 15K under illumination using optical fiber | | Descriptor: | ACETATE ION, CARBON MONOXIDE, FORMIC ACID, ... | | Authors: | Savino, C, Montemiglio, L.C, Ardiccioni, C, Exertier, C, Vallone, B. | | Deposit date: | 2018-11-08 | | Release date: | 2019-09-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Ligand pathways in neuroglobin revealed by low-temperature photodissociation and docking experiments.
Iucrj, 6, 2019
|
|
8QRD
 
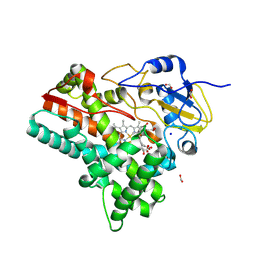 | | OleP in complex with testosterone in high salt crystallization conditions | | Descriptor: | Cytochrome P-450, FORMIC ACID, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Fata, F, Costanzo, A, Freda, I, Gugole, E, Bulfaro, G, Barbizzi, L, Di Renzo, M, Savino, C, Vallone, B, Montemiglio, L.C. | | Deposit date: | 2023-10-06 | | Release date: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | OleP in complex with testosterone in high salt crystallization conditions
To Be Published
|
|
8QYI
 
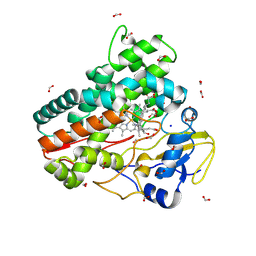 | | OleP in complex with lithocholic acid in high salt crystallization conditions | | Descriptor: | (3beta,5beta,14beta,17alpha)-3-hydroxycholan-24-oic acid, Cytochrome P-450, FORMIC ACID, ... | | Authors: | Fata, F, Costanzo, A, Freda, I, Gugole, E, Bulfaro, G, Barbizzi, L, Di Renzo, M, Savino, C, Vallone, B, Montemiglio, L.C. | | Deposit date: | 2023-10-26 | | Release date: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | OleP in complex with lithocolic acid in high salt crystallization conditions
To Be Published
|
|
1A3O
 
 | | ARTIFICIAL MUTANT (ALPHA Y42H) OF DEOXY HEMOGLOBIN | | Descriptor: | HEMOGLOBIN (ALPHA CHAIN), HEMOGLOBIN (BETA CHAIN), PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Tame, J, Vallone, B. | | Deposit date: | 1998-01-22 | | Release date: | 1998-04-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The structures of deoxy human haemoglobin and the mutant Hb Tyralpha42His at 120 K.
Acta Crystallogr.,Sect.D, 56, 2000
|
|
7ZTH
 
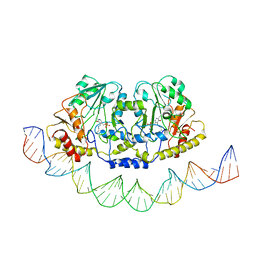 | | Cryo-EM structure of holo-PdxR from Bacillus clausii bound to its target DNA in the open conformation | | Descriptor: | DNA (48-MER), PLP-dependent aminotransferase family protein | | Authors: | Freda, I, Montemiglio, L.C, Tramonti, A, Contestabile, R, Vallone, B, Exertier, C, Savino, C, Chaves Sanjuan, A, Bolognesi, M. | | Deposit date: | 2022-05-10 | | Release date: | 2023-07-05 | | Last modified: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural insights into the DNA recognition mechanism by the bacterial transcription factor PdxR.
Nucleic Acids Res., 51, 2023
|
|
7ZN5
 
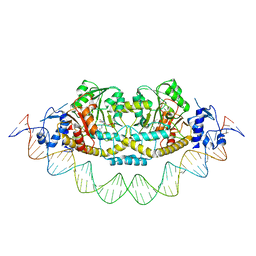 | | Cryo-EM structure of holo-PdxR from Bacillus clausii bound to its target DNA in the closed conformation, C2 symmetry. | | Descriptor: | DNA (48-MER), PLP-dependent aminotransferase family protein | | Authors: | Freda, I, Montemiglio, L.C, Tramonti, A, Contestabile, R, Vallone, B, Exertier, C, Savino, C, Chaves Sanjuan, A, Bolognesi, M. | | Deposit date: | 2022-04-20 | | Release date: | 2023-07-05 | | Last modified: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural insights into the DNA recognition mechanism by the bacterial transcription factor PdxR.
Nucleic Acids Res., 51, 2023
|
|
7ZPA
 
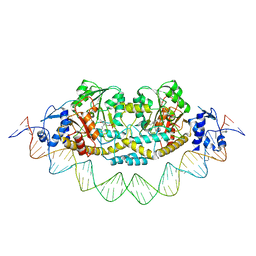 | | Cryo-EM structure of holo-PdxR from Bacillus clausii bound to its target DNA in the closed conformation, C1 symmetry | | Descriptor: | DNA (48-MER), PLP-dependent aminotransferase family protein | | Authors: | Freda, I, Montemiglio, L.C, Tramonti, A, Contestabile, R, Vallone, B, Exertier, C, Savino, C, Chaves Sanjuan, A, Bolognesi, M. | | Deposit date: | 2022-04-27 | | Release date: | 2023-07-05 | | Last modified: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural insights into the DNA recognition mechanism by the bacterial transcription factor PdxR.
Nucleic Acids Res., 51, 2023
|
|
7ZLA
 
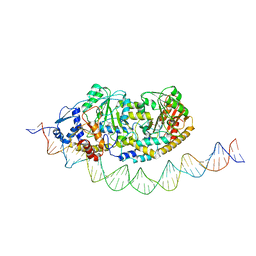 | | Cryo-EM structure of holo-PdxR from Bacillus clausii bound to its target DNA in the half-closed conformation | | Descriptor: | DNA (48-MER), PLP-dependent aminotransferase family protein | | Authors: | Freda, I, Montemiglio, L.C, Tramonti, A, Contestabile, R, Vallone, B, Savino, C, Exertier, C, Bolognesi, M, Chaves Sanjuan, A. | | Deposit date: | 2022-04-14 | | Release date: | 2023-07-05 | | Last modified: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (3.99 Å) | | Cite: | Structural insights into the DNA recognition mechanism by the bacterial transcription factor PdxR.
Nucleic Acids Res., 51, 2023
|
|
1A3N
 
 | | DEOXY HUMAN HEMOGLOBIN | | Descriptor: | HEMOGLOBIN (ALPHA CHAIN), HEMOGLOBIN (BETA CHAIN), PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Tame, J, Vallone, B. | | Deposit date: | 1998-01-22 | | Release date: | 1998-04-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The structures of deoxy human haemoglobin and the mutant Hb Tyralpha42His at 120 K.
Acta Crystallogr.,Sect.D, 56, 2000
|
|
1DXC
 
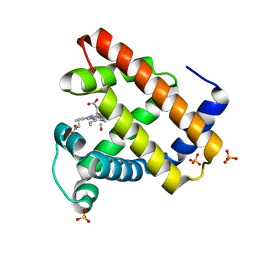 | | CO complex of Myoglobin Mb-YQR at 100K | | Descriptor: | CARBON MONOXIDE, MYOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Brunori, M, Vallone, B, Cutruzzola, F, Travaglini-Allocatelli, C, Berendzen, J, Chu, K, Sweet, R.M, Schlichting, I. | | Deposit date: | 2000-01-03 | | Release date: | 2000-04-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The Role of Cavities in Protein Dynamics: Crystal Structure of a Novel Photolytic Intermediate of Myoglobin
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
