8DOY
 
 | | Crystal structure of SARS-CoV-2 main protease in complex with an inhibitor TKB-198 | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 3C-like proteinase nsp5, ... | | Authors: | Bulut, H, Hayashi, H, Tsuji, K, Kuwata, N, Das, D, Tamamura, H, Mitsuya, H. | | Deposit date: | 2022-07-14 | | Release date: | 2022-08-24 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Potent and biostable inhibitors of the main protease of SARS-CoV-2.
Iscience, 25, 2022
|
|
8UH8
 
 | | Crystal structure of SARS-CoV-2 main protease E166V (Apo structure) | | Descriptor: | ORF1a polyprotein | | Authors: | Bulut, H, Hayashi, H, Kuwata, N, Tsuji, K, Das, D, Tamamura, H, Mitsuya, H. | | Deposit date: | 2023-10-07 | | Release date: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | TKB272, an Orally Available SARS-CoV-2-Mpro Inhibitor Containing 5-Fluorobenzothiazole, Potently Blocks SARS-CoV-2 Replication without Ritonavir
To Be Published
|
|
8UH5
 
 | | Crystal structure of SARS-CoV-2 main protease in complex with an inhibitor TKB-272 | | Descriptor: | (1R,2S,5S)-N-{(1S,2S)-1-(5-fluoro-1,3-benzothiazol-2-yl)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5, DI(HYDROXYETHYL)ETHER | | Authors: | Bulut, H, Hayashi, H, Kuwata, N, Tsuji, K, Das, D, Tamamura, H, Mitsuya, H. | | Deposit date: | 2023-10-06 | | Release date: | 2023-12-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | TKB272, an Orally Available SARS-CoV-2-Mpro Inhibitor Containing 5-Fluorobenzothiazole, Potently Blocks SARS-CoV-2 Replication without Ritonavir
To Be Published
|
|
8UH9
 
 | | Crystal structure of SARS-CoV-2 main protease E166V mutant in complex with an inhibitor TKB-272 | | Descriptor: | (1R,2S,5S)-N-{(1S,2S)-1-(5-fluoro-1,3-benzothiazol-2-yl)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5, DI(HYDROXYETHYL)ETHER | | Authors: | Bulut, H, Hayashi, H, Kuwata, N, Tsuji, K, Das, D, Tamamura, H, Mitsuya, H. | | Deposit date: | 2023-10-07 | | Release date: | 2023-12-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.067 Å) | | Cite: | TKB272, an Orally Available SARS-CoV-2-Mpro Inhibitor Containing 5-Fluorobenzothiazole, Potently Blocks SARS-CoV-2 Replication without Ritonavir
To Be Published
|
|
8DPR
 
 | | Crystal structure of SARS-CoV-2 main protease in complex with inhibitor TKB-248 | | Descriptor: | 2,2,2-trifluoro-N-{(2S)-1-[(1R,2S,5S)-2-({(2S)-1-(4-fluoro-1,3-benzothiazol-2-yl)-1-oxo-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}carbamothioyl)-6,6-dimethyl-3-azabicyclo[3.1.0]hexan-3-yl]-3,3-dimethyl-1-oxobutan-2-yl}acetamide, 3C-like proteinase nsp5, DIMETHYL SULFOXIDE, ... | | Authors: | Bulut, H, Hayashi, H, Tsuji, K, Kuwata, N, Das, D, Tamamura, H, Mitsuya, H. | | Deposit date: | 2022-07-16 | | Release date: | 2022-08-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Identification of SARS-CoV-2 M pro inhibitors containing P1' 4-fluorobenzothiazole moiety highly active against SARS-CoV-2.
Nat Commun, 14, 2023
|
|
8DOX
 
 | | Crystal structure of SARS-CoV-2 main protease in complex with an inhibitor TKB-245 | | Descriptor: | (1R,2S,5S)-N-{(1S,2S)-1-(4-fluoro-1,3-benzothiazol-2-yl)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5, DIMETHYL SULFOXIDE | | Authors: | Bulut, H, Hayashi, H, Tsuji, K, Kuwata, N, Das, D, Tamamura, H, Mitsuya, H. | | Deposit date: | 2022-07-14 | | Release date: | 2022-09-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Identification of SARS-CoV-2 M pro inhibitors containing P1' 4-fluorobenzothiazole moiety highly active against SARS-CoV-2.
Nat Commun, 14, 2023
|
|
1WXZ
 
 | | Crystal structure of adenosine deaminase ligated with a potent inhibitor | | Descriptor: | 1-((1R,2S)-1-{2-[2-(4-CHLOROPHENYL)-1,3-BENZOXAZOL-7-YL]ETHYL}-2-HYDROXYPROPYL)-1H-IMIDAZOLE-4-CARBOXAMIDE, Adenosine deaminase, ZINC ION | | Authors: | Kinoshita, T. | | Deposit date: | 2005-02-02 | | Release date: | 2005-08-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Rational design of non-nucleoside, potent, and orally bioavailable adenosine deaminase inhibitors: predicting enzyme conformational change and metabolism
J.Med.Chem., 48, 2005
|
|
6U85
 
 | |
7MX1
 
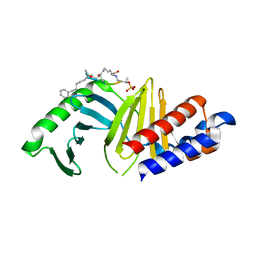 | | PLK-1 polo-box domain in complex with a high affinity macrocycle synthesized using a novel glutamic acid analog | | Descriptor: | ACE-PRO-LEU-ALA-SER-TPO, N-[(4S)-4,5-diamino-5-oxopentyl]-10-phenyldecanamide, Serine/threonine-protein kinase PLK1 | | Authors: | Grant, R.A, Hymel, D, Yaffe, M.B, Burke, T.R. | | Deposit date: | 2021-05-17 | | Release date: | 2022-03-30 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Design and synthesis of a new orthogonally protected glutamic acid analog and its use in the preparation of high affinity polo-like kinase 1 polo-box domain - binding peptide macrocycles.
Org.Biomol.Chem., 19, 2021
|
|
1V7A
 
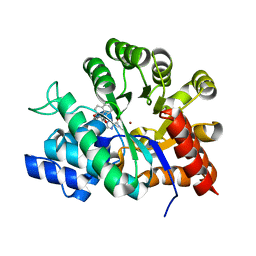 | | Crystal structures of adenosine deaminase complexed with potent inhibitors | | Descriptor: | 1-{(1R,2S)-2-HYDROXY-1-[2-(2-NAPHTHYLOXY)ETHYL]PROPYL}-1H-IMIDAZONE-4-CARBOXAMIDE, ZINC ION, adenosine deaminase | | Authors: | Kinoshita, T. | | Deposit date: | 2003-12-14 | | Release date: | 2004-12-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure-based design and synthesis of non-nucleoside, potent, and orally bioavailable adenosine deaminase inhibitors
J.Med.Chem., 47, 2004
|
|
1V79
 
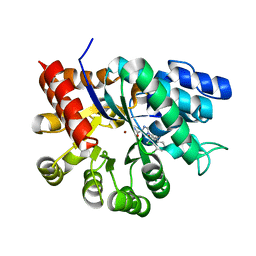 | | Crystal structures of adenosine deaminase complexed with potent inhibitors | | Descriptor: | 1-{(1R,2S)-1-[2-(2,3,-DICHLOROPHENYL)ETHYL]-2-HYDROXYPROPYL}-1H-IMIDAZOLE-4-CARBOXAMIDE, Adenosine deaminase, ZINC ION | | Authors: | Kinoshita, T. | | Deposit date: | 2003-12-14 | | Release date: | 2004-12-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure-based design and synthesis of non-nucleoside, potent, and orally bioavailable adenosine deaminase inhibitors
J.Med.Chem., 47, 2004
|
|
8DCQ
 
 | |
6AX4
 
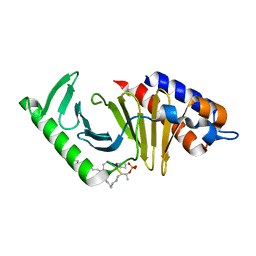 | | Plk-1 polo-box domain in complex with histidine N(tau)-cyclized Macrocycle 5b. | | Descriptor: | AMYLAMINE, Serine/threonine-protein kinase PLK1, histidine N(tau)-cyclized Macrocycle 5b | | Authors: | Grant, R.A, Hymel, D, Yaffe, M.B, Burke, T.R. | | Deposit date: | 2017-09-06 | | Release date: | 2018-09-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Histidine N( tau )-cyclized macrocycles as a new genre of polo-like kinase 1 polo-box domain-binding inhibitors.
Bioorg. Med. Chem. Lett., 28, 2018
|
|
2E1W
 
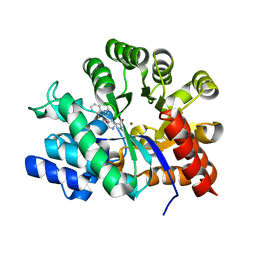 | | Crystal structure of adenosine deaminase complexed with potent inhibitors | | Descriptor: | 1-{(1R,2S)-2-HYDROXY-1-[2-(1-NAPHTHYL)ETHYL]PROPYL}-1H-IMIDAZOLE-4-CARBOXAMIDE, Adenosine deaminase, ZINC ION | | Authors: | Kinoshita, T. | | Deposit date: | 2006-10-30 | | Release date: | 2006-11-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure-Based Design and Synthesis of Non-Nucleoside, Potent, and Orally Bioavailable Adenosine Deaminase Inhibitors
J.Med.Chem., 47, 2004
|
|
2DYS
 
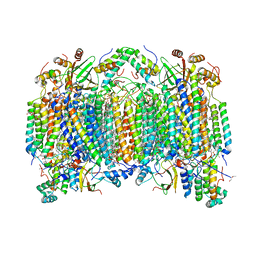 | | Bovine heart cytochrome C oxidase modified by DCCD | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Shinzawa-Itoh, K, Aoyama, H, Muramoto, K, Kurauchi, T, Mizushima, T, Yamashita, E, Tsukihara, T, Yoshikawa, S. | | Deposit date: | 2006-09-16 | | Release date: | 2007-04-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structures and physiological roles of 13 integral lipids of bovine heart cytochrome c oxidase
Embo J., 26, 2007
|
|
2EFL
 
 | | Crystal structure of the EFC domain of formin-binding protein 17 | | Descriptor: | Formin-binding protein 1 | | Authors: | Shimada, A, Niwa, H, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-02-23 | | Release date: | 2007-05-29 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Curved EFC/F-BAR-Domain Dimers Are Joined End to End into a Filament for Membrane Invagination in Endocytosis
Cell(Cambridge,Mass.), 129, 2007
|
|
2DYR
 
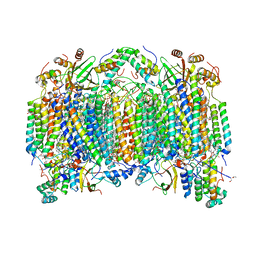 | | Bovine heart cytochrome C oxidase at the fully oxidized state | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Shinzawa-Itoh, K, Aoyama, H, Muramoto, K, Kurauchi, T, Mizushima, T, Yamashita, E, Tsukihara, T, Yoshikawa, S. | | Deposit date: | 2006-09-16 | | Release date: | 2007-04-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures and physiological roles of 13 integral lipids of bovine heart cytochrome c oxidase
Embo J., 26, 2007
|
|
2EFK
 
 | | Crystal structure of the EFC domain of Cdc42-interacting protein 4 | | Descriptor: | Cdc42-interacting protein 4 | | Authors: | Shimada, A, Niwa, H, Chen, L, Liu, Z.-J, Wang, B.-C, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-02-23 | | Release date: | 2007-05-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Curved EFC/F-BAR-Domain Dimers Are Joined End to End into a Filament for Membrane Invagination in Endocytosis
Cell(Cambridge,Mass.), 129, 2007
|
|
