1XA4
 
 | | Crystal structure of CaiB, a type III CoA transferase in carnitine metabolism | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, COENZYME A, Crotonobetainyl-CoA:carnitine CoA-transferase, ... | | Authors: | Stenmark, P, Gurmu, D, Nordlund, P. | | Deposit date: | 2004-08-25 | | Release date: | 2004-11-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of CaiB, a Type-III CoA Transferase in Carnitine Metabolism
Biochemistry, 43, 2004
|
|
1XA3
 
 | | Crystal structure of CaiB, a type III CoA transferase in carnitine metabolism | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Crotonobetainyl-CoA:carnitine CoA-transferase, SULFATE ION | | Authors: | Stenmark, P, Gurmu, D, Nordlund, P, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2004-08-25 | | Release date: | 2004-11-16 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structure of CaiB, a Type-III CoA Transferase in Carnitine Metabolism
Biochemistry, 43, 2004
|
|
2CAR
 
 | | Crystal Structure Of Human Inosine Triphosphatase | | Descriptor: | INOSINE TRIPHOSPHATE PYROPHOSPHATASE | | Authors: | Stenmark, P, Kursula, P, Arrowsmith, C, Berglund, H, Edwards, A, Ehn, M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Hallberg, B.M, Hogbom, M, Holmberg Schiavone, L, Kotenyova, T, Nilsson-Ehle, P, Nyman, T, Ogg, D, Persson, C, Sagemark, J, Schuler, H, Sundstrom, M, Thorsell, A.G, van den Berg, S, Weigelt, J, Nordlund, P. | | Deposit date: | 2005-12-22 | | Release date: | 2006-01-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.09 Å) | | Cite: | Crystal Structure of Human Inosine Triphosphatase. Substrate Binding and Implication of the Inosine Triphosphatase Deficiency Mutation P32T.
J.Biol.Chem., 282, 2007
|
|
2VD6
 
 | | Human adenylosuccinate lyase in complex with its substrate N6-(1,2- Dicarboxyethyl)-AMP, and its products AMP and fumarate. | | Descriptor: | 2-[9-(3,4-DIHYDROXY-5-PHOSPHONOOXYMETHYL-TETRAHYDRO-FURAN-2-YL)-9H-PURIN-6-YLAMINO]-SUCCINIC ACID, ADENOSINE MONOPHOSPHATE, ADENYLOSUCCINATE LYASE, ... | | Authors: | Stenmark, P, Moche, M, Arrowsmith, C, Berglund, H, Busam, R, Collins, R, Dahlgren, L.G, Edwards, A, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Hallberg, B.M, Holmberg-schiavone, L, Johansson, I, Kallas, A, Karlberg, T, Kotenyova, T, Lehtio, L, Nilsson, M, Nyman, T, Ogg, D, Persson, C, Sagemark, J, Sundstrom, M, Thorsell, A.G, Tresaugues, L, van den Berg, S, Weigelt, J, Welin, M, Nordlund, P, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-09-30 | | Release date: | 2007-10-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Human Adenylosuccinate Lyase in Complex with its Substrate N6-(1,2-Dicarboxyethyl)-AMP, and its Products AMP and Fumarate.
To be Published
|
|
2J4E
 
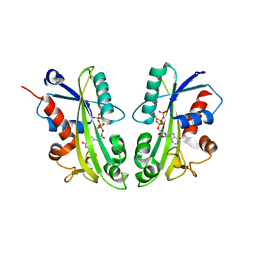 | | THE ITP COMPLEX OF HUMAN INOSINE TRIPHOSPHATASE | | Descriptor: | INOSINE 5'-TRIPHOSPHATE, INOSINE TRIPHOSPHATE PYROPHOSPHATASE, INOSINIC ACID, ... | | Authors: | Stenmark, P, Kursula, P, Arrowsmith, C, Berglund, H, Busam, R, Collins, R, Edwards, A, Ehn, M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Hallberg, B.M, Holmbergschiavone, L, Hogbom, M, Kotenyova, T, Landry, R, Loppnau, P, Magnusdottir, A, Nilsson-Ehle, P, Nyman, T, Ogg, D, Persson, C, Sagemark, J, Sundstrom, M, Uppenberg, J, Thorsell, A.G, Schuler, H, Van Den Berg, S, Wallden, K, Weigelt, J, Nordlund, P. | | Deposit date: | 2006-08-29 | | Release date: | 2006-09-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of Human Inosine Triphosphatase: Substrate Binding and Implication of the Inosine Triphosphatase Deficiency Mutation P32T.
J.Biol.Chem., 282, 2007
|
|
2J91
 
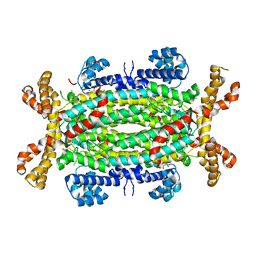 | | Crystal structure of Human Adenylosuccinate Lyase in complex with AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, ADENYLOSUCCINATE LYASE, CHLORIDE ION, ... | | Authors: | Stenmark, P, Moche, M, Arrowsmith, C, Berglund, H, Busam, R, Collins, R, Edwards, A, Ericsson, U.B, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Hallberg, B.M, Holmberg Schiavone, L, Hogbom, M, Johansson, I, Karlberg, T, Kosinska, U, Kotenyova, T, Magnusdottir, A, Nilsson, M.E, Nilsson-Ehle, P, Nyman, T, Ogg, D, Persson, C, Sagemark, J, Sundstrom, M, Uppenberg, J, Uppsten, M, Thorsell, A.G, van Den Berg, S, Wallden, K, Weigelt, J, Nordlund, P. | | Deposit date: | 2006-11-01 | | Release date: | 2006-11-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of Human Adenylosuccinate Lyase
To be Published
|
|
2G6V
 
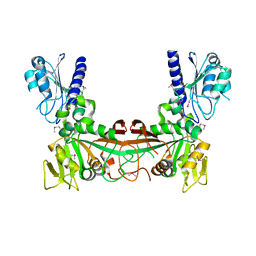 | | The crystal structure of ribD from Escherichia coli | | Descriptor: | Riboflavin biosynthesis protein ribD | | Authors: | Stenmark, P, Moche, M, Gurmu, D, Nordlund, P, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2006-02-25 | | Release date: | 2007-02-06 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The Crystal Structure of the Bifunctional Deaminase/Reductase RibD of the Riboflavin Biosynthetic Pathway in Escherichia coli: Implications for the Reductive Mechanism.
J.Mol.Biol., 373, 2007
|
|
2VU9
 
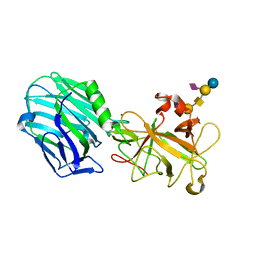 | | CRYSTAL STRUCTURE OF BOTULINUM NEUROTOXIN SEROTYPE A BINDING DOMAIN IN COMPLEX WITH GT1B | | Descriptor: | BOTULINUM NEUROTOXIN A HEAVY CHAIN, MAGNESIUM ION, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-galactopyranose-(1-4)-[N-acetyl-alpha-neuraminic acid-(2-3)]beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Stenmark, P, Dupuy, J, Stevens, R.C. | | Deposit date: | 2008-05-22 | | Release date: | 2008-08-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure of Botulinum Neurotoxin Type a in Complex with the Cell Surface Co-Receptor Gt1B- Insight Into the Toxin-Neuron Interaction.
Plos Pathog., 4, 2008
|
|
2VUA
 
 | |
2VXR
 
 | |
2BZN
 
 | | Crystal structure of human guanosine monophosphate reductase 2 GMPR2 in complex with IMP | | Descriptor: | GMP REDUCTASE 2, INOSINIC ACID | | Authors: | Stenmark, P, Kursula, P, Arrowsmith, C, Berglund, H, Edwards, A, Ehn, M, Graslund, S, Hammarstrom, M, Hallberg, B.M, Kotenyova, T, Nilsson-Ehle, P, Nordlund, P, Ogg, D, Persson, C, Sagemark, J, Schuler, H, Sundstrom, M, Thorsell, A, Weigelt, J. | | Deposit date: | 2005-08-19 | | Release date: | 2005-09-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Cofactor Mobility Determines Reaction Outcome in the Impdh and Gmpr (Beta-Alpha)(8) Barrel Enzymes.
Nat.Chem.Biol., 7, 2011
|
|
2VO1
 
 | | CRYSTAL STRUCTURE OF THE SYNTHETASE DOMAIN OF HUMAN CTP SYNTHETASE | | Descriptor: | CTP SYNTHASE 1, SULFATE ION | | Authors: | Stenmark, P, Kursula, P, Arrowsmith, C, Berglund, H, Edwards, A, Ehn, M, Flodin, S, Graslund, S, Hammarstrom, M, Hallberg, B.M, Holmberg-Schiavone, L, Kotenyoa, T, Moche, M, Nilsson-Ehle, P, Ogg, D, Persson, C, Sagemark, J, Schuler, H, Sundstrom, M, Thorsell, A.G, Van Den Berg, S, Weigelt, J, Nordlund, P. | | Deposit date: | 2008-02-08 | | Release date: | 2008-03-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the Synthetase Domain of Human Ctp Synthetase, a Target for Anticancer Therapy.
Acta Crystallogr.,Sect.F, 62, 2006
|
|
2CL3
 
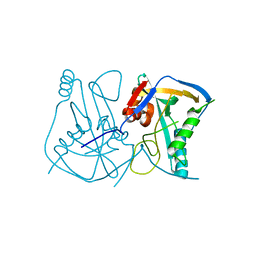 | | Crystal structure of human Cleavage and Polyadenylation Specificity Factor 5 (CPSF5) | | Descriptor: | CLEAVAGE AND POLYADENYLATION SPECIFICITY FACTOR 5 | | Authors: | Stenmark, P, Hogbom, M, Arrowsmith, C, Berglund, H, Collins, R, Edwards, A, Ehn, M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Hallberg, B.M, Holmberg Schiavone, L, Kotenyova, T, Magnusdottir, A, Nilsson-Ehle, P, Nyman, T, Ogg, D, Persson, C, Sagemark, J, Sundstrom, M, Thorsell, A.G, Van Den Berg, S, Wallden, K, Weigelt, J, Nordlund, P. | | Deposit date: | 2006-04-25 | | Release date: | 2006-05-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Crystal Structure of Human Cleavage and Polyadenylation Specific Factor-5 Reveals a Dimeric Nudix Protein with a Conserved Catalytic Site.
Proteins, 73, 2008
|
|
2GSE
 
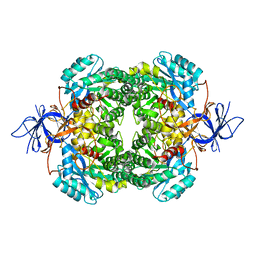 | | Crystal Structure of Human Dihydropyrimidinease-like 2 | | Descriptor: | CALCIUM ION, Dihydropyrimidinase-related protein 2 | | Authors: | Ogg, D, Stenmark, P, Arrowsmith, C, Berglund, H, Collins, R, Edwards, A, Ehn, M, Flodin, S, Flores, A, Graslund, S, Hallberg, B.M, Hammarstrom, M, Kotenyova, T, Kursula, P, Nilsson-Ehle, P, Nyman, T, Persson, C, Sagemark, J, Sundstrom, M, Holmberg-Schiavone, L, Thorsell, A.G, Uppenberg, J, Van Den Berg, S, Weigelt, J, Nordlund, P, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-04-26 | | Release date: | 2006-05-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The structure of human collapsin response mediator protein 2, a regulator of axonal growth.
J.Neurochem., 101, 2007
|
|
2OBC
 
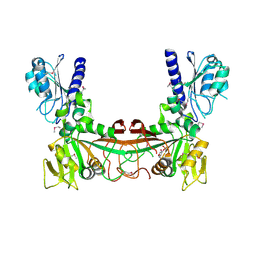 | | The crystal structure of RibD from Escherichia coli in complex with a substrate analogue, ribose 5-phosphate (beta form), bound to the active site of the reductase domain | | Descriptor: | 5-O-phosphono-beta-D-ribofuranose, Riboflavin biosynthesis protein ribD | | Authors: | Moche, M, Stenmark, P, Gurmu, D, Nordlund, P, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2006-12-18 | | Release date: | 2007-02-13 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The crystal structure of the bifunctional deaminase/reductase RibD of the riboflavin biosynthetic pathway in Escherichia coli: implications for the reductive mechanism.
J.Mol.Biol., 373, 2007
|
|
2O7P
 
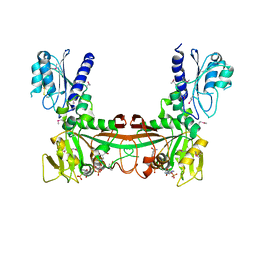 | | The crystal structure of RibD from Escherichia coli in complex with the oxidised NADP+ cofactor in the active site of the reductase domain | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Riboflavin biosynthesis protein ribD | | Authors: | Moche, M, Stenmark, P, Gurmu, D, Nordlund, P, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2006-12-11 | | Release date: | 2007-02-13 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The crystal structure of the bifunctional deaminase/reductase RibD of the riboflavin biosynthetic pathway in Escherichia coli: implications for the reductive mechanism.
J.Mol.Biol., 373, 2007
|
|
2J67
 
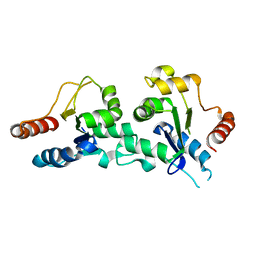 | | The TIR domain of human Toll-Like Receptor 10 (TLR10) | | Descriptor: | TOLL LIKE RECEPTOR 10 | | Authors: | Stenmark, P, Ogg, D, Arrowsmith, C, Berglund, H, Busam, R, Collins, R, Edwards, A, Ericsson, U.B, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Hallberg, B.M, Holmberg Schiavone, L, Hogbom, M, Johansson, I, Karlberg, T, Kotenyova, T, Magnusdottir, A, Nilsson, M.E, Nilsson-Ehle, P, Nyman, T, Persson, C, Sagemark, J, Sundstrom, M, Uppenberg, J, Thorsell, A.G, Van Den Berg, S, Wallden, K, Weigelt, J, Welin, M, Nordlund, P. | | Deposit date: | 2006-09-26 | | Release date: | 2006-09-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Crystal Structure of the Human Toll-Like Receptor 10 Cytoplasmic Domain Reveals a Putative Signaling Dimer.
J.Biol.Chem., 283, 2008
|
|
4N1T
 
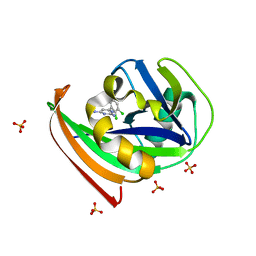 | | Structure of human MTH1 in complex with TH287 | | Descriptor: | 6-(2,3-dichlorophenyl)-N~4~-methylpyrimidine-2,4-diamine, 7,8-dihydro-8-oxoguanine triphosphatase, SULFATE ION | | Authors: | Berntsson, R.P.-A, Jemth, A, Gustafsson, R, Svensson, L.M, Helleday, T, Stenmark, P. | | Deposit date: | 2013-10-04 | | Release date: | 2014-04-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | MTH1 inhibition eradicates cancer by preventing sanitation of the dNTP pool.
Nature, 508, 2014
|
|
6ZVN
 
 | |
5C6K
 
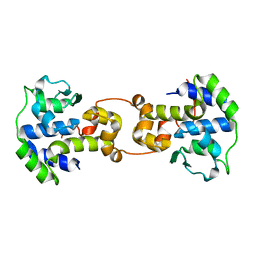 | | Bacteriophage P2 integrase catalytic domain | | Descriptor: | Integrase | | Authors: | Skaar, K, Claesson, M, Odegrip, R, Eriksson, J, Hogbom, M, Haggard-Ljungquist, E, Stenmark, P. | | Deposit date: | 2015-06-23 | | Release date: | 2015-10-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the bacteriophage P2 integrase catalytic domain.
Febs Lett., 589, 2015
|
|
7ZC7
 
 | | Structure of the mouse 8-oxoguanine DNA Glycosylase mOGG1 in complex with ligand TH012941 | | Descriptor: | 2-[4-(3,5-dimethylpyrazol-1-yl)phenyl]-~{N}-(4,5,6,7-tetrahydro-1,2-benzoxazol-3-yl)ethanamide, N-glycosylase/DNA lyase, NICKEL (II) ION | | Authors: | Davies, J.R, Scaletti, E, Stenmark, P. | | Deposit date: | 2022-03-25 | | Release date: | 2023-07-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the mouse 8-oxoguanine DNA Glycosylase mOGG1 in complex with ligand TH012941
To Be Published
|
|
8BVX
 
 | |
8BYP
 
 | |
6HOX
 
 | |
8C5H
 
 | |
