1YET
 
 | |
1YER
 
 | |
1VCB
 
 | | THE VHL-ELONGINC-ELONGINB STRUCTURE | | Descriptor: | PROTEIN (ELONGIN B), PROTEIN (ELONGIN C), PROTEIN (VHL) | | Authors: | Stebbins, C.E, Kaelin, W.G, Pavletich, N.P. | | Deposit date: | 1999-03-13 | | Release date: | 1999-04-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of the VHL-ElonginC-ElonginB complex: implications for VHL tumor suppressor function.
Science, 284, 1999
|
|
1YES
 
 | |
1JYO
 
 | |
1G4W
 
 | |
1G4U
 
 | |
6ELC
 
 | | Crystal Structure of O-linked Glycosylated VSG3 | | Descriptor: | Variant surface glycoprotein, alpha-D-glucopyranose, alpha-D-mannopyranose-(1-6)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Stebbins, C.E. | | Deposit date: | 2017-09-28 | | Release date: | 2018-07-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | African trypanosomes evade immune clearance by O-glycosylation of the VSG surface coat.
Nat Microbiol, 3, 2018
|
|
3EFY
 
 | |
1GRJ
 
 | |
3LW9
 
 | |
3IEC
 
 | |
1Q5Z
 
 | | Crystal Structure of the C-terminal Actin Binding Domain of Salmonella Invasion Protein A (SipA) | | Descriptor: | SipA | | Authors: | Stebbins, C.E, Lilic, M, Galkin, V.E, Orlova, A, VanLoock, M.S, Egelman, E.H. | | Deposit date: | 2003-08-11 | | Release date: | 2003-10-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Salmonella SipA polymerizes actin by stapling filaments with nonglobular protein arms.
Science, 301, 2003
|
|
4G29
 
 | |
4GF3
 
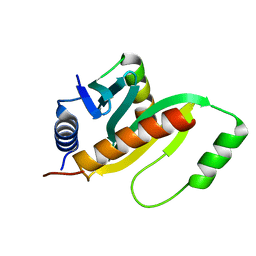 | | Structure of a SycH-YopH Chaperone-Effector Complex | | Descriptor: | Putative yopH targeting protein, Tyrosine-protein phosphatase yopH | | Authors: | Stebbins, C.E, Vujanac, M. | | Deposit date: | 2012-08-02 | | Release date: | 2013-03-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Context-dependent protein folding of a virulence peptide in the bacterial and host environments: structure of an SycH-YopH chaperone-effector complex.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4G2B
 
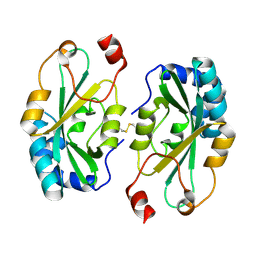 | |
4G6T
 
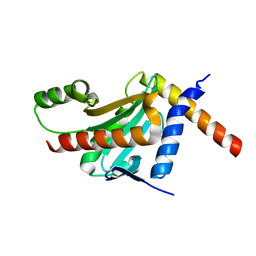 | | Structure of the HopA1-SchA Chaperone-Effector Complex | | Descriptor: | Type III chaperone protein ShcA, Type III effector HopA1 | | Authors: | Stebbins, C.E, Janjusevic, R, Quezada, C.M. | | Deposit date: | 2012-07-19 | | Release date: | 2013-06-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Structure of the HopA1(21-102)-ShcA chaperone-effector complex of Pseudomonas syringae reveals conservation of a virulence factor binding motif from animal to plant pathogens.
J.Bacteriol., 195, 2013
|
|
4IRV
 
 | |
6Z8G
 
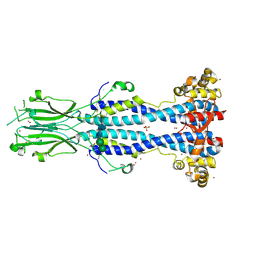 | | Crystal structure of VSG13 soaked in 0.5 M used to phase VSG13 to solve the structure. | | Descriptor: | BROMIDE ION, SULFATE ION, Variant surface glycoprotein MITat 1.13, ... | | Authors: | Stebbins, C.E, Hempelmann, A, Van Straaten, M, Zeelen, J. | | Deposit date: | 2020-06-02 | | Release date: | 2021-03-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Structure of trypanosome coat protein VSGsur and function in suramin resistance.
Nat Microbiol, 6, 2021
|
|
6Z8H
 
 | | Crystal structure of Variant Surface Glycoprotein VSG13 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, SULFATE ION, Variant surface glycoprotein MITat 1.13, ... | | Authors: | Stebbins, C.E, Hempelmann, A, Van Straaten, M, Zeelen, J. | | Deposit date: | 2020-06-02 | | Release date: | 2021-03-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Structure of trypanosome coat protein VSGsur and function in suramin resistance.
Nat Microbiol, 6, 2021
|
|
7AQX
 
 | |
7AQY
 
 | |
7AR0
 
 | |
7AQZ
 
 | | Co-Crystal Structure of Variant Surface Glycoprotein VSG2 in complex with Nanobody VSG2(NB14) | | Descriptor: | CITRIC ACID, Nanobody VSG2(NB14), SODIUM ION, ... | | Authors: | Stebbins, C.E, Hempelmann, A, VanStraaten, M. | | Deposit date: | 2020-10-23 | | Release date: | 2021-11-03 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Nanobody-mediated macromolecular crowding induces membrane fission and remodeling in the African trypanosome.
Cell Rep, 37, 2021
|
|
2GWK
 
 | |
