2XTN
 
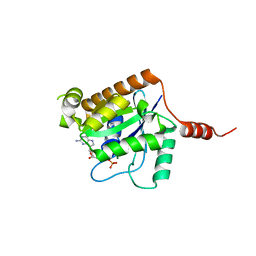 | | Crystal structure of GTP-bound human GIMAP2, amino acid residues 1- 234 | | Descriptor: | GTPASE IMAP FAMILY MEMBER 2, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION | | Authors: | Schwefel, D, Froehlich, C, Daumke, O. | | Deposit date: | 2010-10-11 | | Release date: | 2010-10-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Basis of Oligomerization in Septin-Like Gtpase of Immunity-Associated Protein 2 (Gimap2)
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
2XTP
 
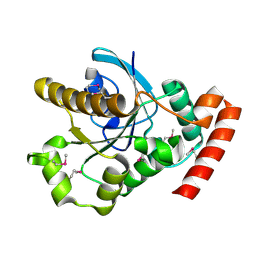 | |
2XTM
 
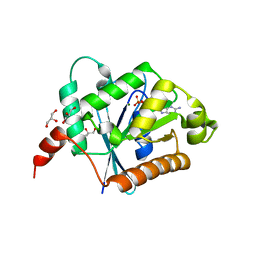 | | Crystal structure of GDP-bound human GIMAP2, amino acid residues 1- 234 | | Descriptor: | GLYCEROL, GTPASE IMAP FAMILY MEMBER 2, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Schwefel, D, Froehlich, C, Daumke, O. | | Deposit date: | 2010-10-11 | | Release date: | 2010-10-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Basis of Oligomerization in Septin-Like Gtpase of Immunity-Associated Protein 2 (Gimap2)
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
2XTO
 
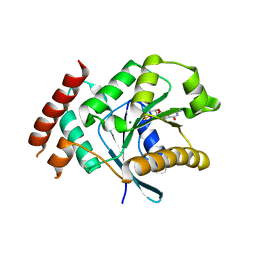 | | Crystal structure of GDP-bound human GIMAP2, amino acid residues 21- 260 | | Descriptor: | GTPASE IMAP FAMILY MEMBER 2, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION | | Authors: | Schwefel, D, Froehlich, C, Daumke, O. | | Deposit date: | 2010-10-11 | | Release date: | 2010-10-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Basis of Oligomerization in Septin-Like Gtpase of Immunity-Associated Protein 2 (Gimap2)
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
6Z3E
 
 | |
3ZJC
 
 | |
2UVO
 
 | | High Resolution Crystal Structure of Wheat Germ Agglutinin in Complex with N-Acetyl-D-Glucosamine | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, AGGLUTININ ISOLECTIN 1, ... | | Authors: | Schwefel, D, Wittmann, V, Diederichs, K, Welte, W. | | Deposit date: | 2007-03-13 | | Release date: | 2008-05-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural Basis of Multivalent Binding to Wheat Germ Agglutinin.
J.Am.Chem.Soc., 132, 2010
|
|
2X52
 
 | | CRYSTAL STRUCTURE OF WHEAT GERM AGGLUTININ ISOLECTIN 3 IN COMPLEX WITH A SYNTHETIC DIVALENT CARBOHYDRATE LIGAND | | Descriptor: | AGGLUTININ ISOLECTIN 3, BIS-(2-ACETAMIDO-2-DEOXY-ALPHA-D-GLUCOPYRANOSYLOXYCARBONYL)-4,7,10-TRIOXA-1,13-TRIDECANEDIAMINE, GLYCEROL | | Authors: | Schwefel, D, Maierhofer, C, Wittmann, V, Diederichs, K, Welte, W. | | Deposit date: | 2010-02-05 | | Release date: | 2010-02-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Basis of Multivalent Binding to Wheat Germ Agglutinin.
J.Am.Chem.Soc., 132, 2010
|
|
2X3T
 
 | | Glutaraldehyde-crosslinked wheat germ agglutinin isolectin 1 crystal soaked with a synthetic glycopeptide | | Descriptor: | 2-acetamido-1-O-carbamoyl-2-deoxy-alpha-D-glucopyranose, AGGLUTININ ISOLECTIN 1, D-ALPHA-AMINOBUTYRIC ACID, ... | | Authors: | Schwefel, D, Maierhofer, C, Wittmann, V, Diederichs, K, Welte, W. | | Deposit date: | 2010-01-26 | | Release date: | 2010-06-30 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.749 Å) | | Cite: | Structural Basis of Multivalent Binding to Wheat Germ Agglutinin.
J.Am.Chem.Soc., 132, 2010
|
|
4CC9
 
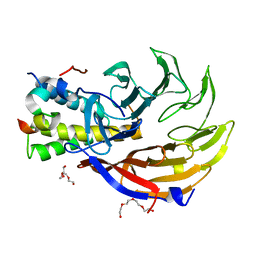 | | Crystal structure of human SAMHD1 (amino acid residues 582-626) bound to Vpx isolated from sooty mangabey and human DCAF1 (amino acid residues 1058-1396) | | Descriptor: | DEOXYNUCLEOSIDE TRIPHOSPHATE TRIPHOSPHOHYDROLASE SAMHD1, PROTEIN VPRBP, PROTEIN VPX, ... | | Authors: | Schwefel, D, Groom, H.C.T, Boucherit, V.C, Christodoulou, E, Walker, P.A, Stoye, J.P, Bishop, K.N, Taylor, I.A. | | Deposit date: | 2013-10-19 | | Release date: | 2013-12-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.473 Å) | | Cite: | Structural Basis of Lentiviral Subversion of a Cellular Protein Degradation Pathway.
Nature, 505, 2014
|
|
4AML
 
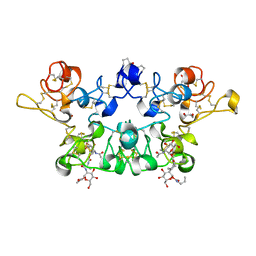 | | CRYSTAL STRUCTURE OF WHEAT GERM AGGLUTININ ISOLECTIN 1 IN COMPLEX WITH GLYCOSYLURETHAN | | Descriptor: | 2-acetamido-2-deoxy-1-O-(propylcarbamoyl)-alpha-D-glucopyranose, AGGLUTININ ISOLECTIN 1, GLYCEROL | | Authors: | Schwefel, D, Maierhofer, C, Beck, J.G, Seeberger, S, Diederichs, K, Moeller, H.M, Welte, W, Wittmann, V. | | Deposit date: | 2012-03-12 | | Release date: | 2012-04-04 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural Basis of Multivalent Binding to Wheat Germ Agglutinin.
J.Am.Chem.Soc., 132, 2010
|
|
5AJA
 
 | | Crystal structure of mandrill SAMHD1 (amino acid residues 1-114) bound to Vpx isolated from mandrill and human DCAF1 (amino acid residues 1058-1396) | | Descriptor: | PROTEIN VPRBP, SAM DOMAIN AND HD DOMAIN-CONTAINING PROTEIN, VPX PROTEIN, ... | | Authors: | Schwefel, D, Boucherit, V.C, Christodoulou, E, Walker, P.A, Stoye, J.P, Bishop, K.N, Taylor, I.A. | | Deposit date: | 2015-02-20 | | Release date: | 2015-04-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.649 Å) | | Cite: | Molecular Determinants for Recognition of Divergent Samhd1 Proteins by the Lentiviral Accessory Protein Vpx.
Cell Host Microbe., 17, 2015
|
|
8BGG
 
 | | Cryo-EM structure of SARS-CoV-2 spike (Omicron BA.1 variant) in complex with nanobody W25 (map 5, focus refinement on RBD, W25 and adjacent NTD) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Nanobody W25, Spike glycoprotein, ... | | Authors: | Modhiran, N, Lauer, S, Spahn, C.M.T, Watterson, D, Schwefel, D. | | Deposit date: | 2022-10-27 | | Release date: | 2023-03-08 | | Method: | ELECTRON MICROSCOPY (6.04 Å) | | Cite: | Cryo-EM structure of SARS-CoV-2 spike (Omicron BA.1 variant) in complex with nanobody W25 (map 5, focus refinement on RBD, W25 and adjacent NTD)
To Be Published
|
|
8BEV
 
 | | Cryo-EM structure of SARS-CoV-2 spike (HexaPro variant) in complex with nanobody W25 (map 3, focus refinement on RBD, W25 and adjacent NTD) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Lauer, S, Spahn, C.M.T, Schwefel, D. | | Deposit date: | 2022-10-21 | | Release date: | 2023-03-08 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (5.92 Å) | | Cite: | Cryo-EM structure of SARS-CoV-2 spike (HexaPro variant) in complex with nanobody W25 (map 3, focus refinement on RBD, W25 and adjacent NTD)
To Be Published
|
|
5AO1
 
 | |
5AO3
 
 | |
5AO0
 
 | |
5AO2
 
 | |
6ZX9
 
 | | Crystal structure of SIV Vpr,fused to T4 lysozyme, isolated from moustached monkey, bound to human DDB1 and human DCAF1 (amino acid residues 1046-1396) | | Descriptor: | DDB1- and CUL4-associated factor 1, DNA damage-binding protein 1, GLYCEROL, ... | | Authors: | Schwefel, D, Banchenko, S. | | Deposit date: | 2020-07-29 | | Release date: | 2021-07-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.519729 Å) | | Cite: | Structural insights into Cullin4-RING ubiquitin ligase remodelling by Vpr from simian immunodeficiency viruses.
Plos Pathog., 17, 2021
|
|
6ZUE
 
 | |
7ZNN
 
 | | Cryo-EM structure of RCMV-E E27 bound to human DDB1 (deltaBPB) and full-length rat STAT2 | | Descriptor: | B27a, DNA damage-binding protein 1, Signal transducer and activator of transcription, ... | | Authors: | Lauer, S, Spahn, C.M.T, Schwefel, D. | | Deposit date: | 2022-04-21 | | Release date: | 2022-11-09 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Structural mechanism of CRL4-instructed STAT2 degradation via a novel cytomegaloviral DCAF receptor.
Embo J., 42, 2023
|
|
7ZN7
 
 | | Cryo-EM structure of RCMV-E E27 bound to human DDB1 (deltaBPB) and rat STAT2 CCD | | Descriptor: | B27a, DNA damage-binding protein 1, Signal transducer and activator of transcription, ... | | Authors: | Lauer, S, Spahn, C.M.T, Schwefel, D. | | Deposit date: | 2022-04-20 | | Release date: | 2022-11-09 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.78 Å) | | Cite: | Structural mechanism of CRL4-instructed STAT2 degradation via a novel cytomegaloviral DCAF receptor.
Embo J., 42, 2023
|
|
4FVF
 
 | |
4FVG
 
 | |
4FVJ
 
 | |
