4UP5
 
 | | Crystal structure of the Pygo2 PHD finger in complex with the B9L HD1 domain and a chemical fragment | | Descriptor: | 6-methoxy-1,3-benzothiazol-2-amine, PYGOPUS HOMOLOG 2, B-CELL CLL/LYMPHOMA 9-LIKE PROTEIN, ... | | Authors: | Miller, T.C.R, Fiedler, M, Rutherford, T.J, Birchall, K, Chugh, J, Bienz, M. | | Deposit date: | 2014-06-12 | | Release date: | 2014-11-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Competitive Binding of a Benzimidazole to the Histone-Binding Pocket of the Pygo Phd Finger.
Acs Chem.Biol., 9, 2014
|
|
4UP0
 
 | | Ternary crystal structure of the Pygo2 PHD finger in complex with the B9L HD1 domain and a H3K4me2 peptide | | Descriptor: | HISTONE H3.1, PYGOPUS HOMOLOG 2, B-CELL CLL/LYMPHOMA 9-LIKE PROTEIN, ... | | Authors: | Miller, T.C.R, Fiedler, M, Rutherford, T.J, Birchall, K, Chugh, J, Bienz, M. | | Deposit date: | 2014-06-11 | | Release date: | 2014-11-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Competitive Binding of a Benzimidazole to the Histone-Binding Pocket of the Pygo Phd Finger.
Acs Chem.Biol., 9, 2014
|
|
7NWJ
 
 | |
2FEJ
 
 | | Solution structure of human p53 DNA binding domain. | | Descriptor: | Cellular tumor antigen p53, ZINC ION | | Authors: | Perez-Canadillas, J.M, Tidow, H, Freund, S.M, Rutherford, T.J, Ang, H.C, Fersht, A.R. | | Deposit date: | 2005-12-16 | | Release date: | 2006-01-31 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of p53 core domain: Structural basis for its instability
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
3ZPV
 
 | | Crystal structure of Drosophila Pygo PHD finger in complex with Legless HD1 domain | | Descriptor: | PROTEIN BCL9 HOMOLOG, PROTEIN PYGOPUS, ZINC ION | | Authors: | Miller, T.C.R, Mieszczanek, J, Sanchez-Barrena, M.J, Rutherford, T.J, Fiedler, M, Bienz, M. | | Deposit date: | 2013-03-02 | | Release date: | 2013-10-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Evolutionary Adaptation of the Fly Pygo Phd Finger Towards Recognizing Histone H3 Tail Methylated at Arginine 2
Structure, 21, 2013
|
|
2UWQ
 
 | |
2WQG
 
 | | SAP domain from Tho1: L31W (fluorophore) mutant | | Descriptor: | PROTEIN THO1 | | Authors: | Dodson, C.A, Ferguson, N, Rutherford, T.J, Johnson, C.M, Fersht, A.R. | | Deposit date: | 2009-08-21 | | Release date: | 2010-02-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Engineering a Two-Helix Bundle Protein for Folding Studies.
Protein Eng.Des.Sel., 23, 2010
|
|
2XB1
 
 | | Crystal structure of the human Pygo2 PHD finger in complex with the B9L HD1 domain | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, PYGOPUS HOMOLOG 2, ... | | Authors: | Miller, T.C, Rutherford, T.J, Johnson, C.M, Fiedler, M, Bienz, M. | | Deposit date: | 2010-04-01 | | Release date: | 2010-07-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Allosteric Remodeling of the Histone H3 Binding Pocket in the Pygo2 Phd Finger Triggered by its Binding to the B9L/Bcl9 Co-Factor.
J.Mol.Biol., 401, 2010
|
|
2VY4
 
 | | U11-48K CHHC ZN-FINGER DOMAIN | | Descriptor: | U11/U12 SMALL NUCLEAR RIBONUCLEOPROTEIN 48 KDA PROTEIN, ZINC ION | | Authors: | Tidow, H, Andreeva, A, Rutherford, T.J, Fersht, A.R. | | Deposit date: | 2008-07-17 | | Release date: | 2009-02-17 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the U11-48K CHHC zinc-finger domain that specifically binds the 5' splice site of U12-type introns.
Structure, 17, 2009
|
|
2VY5
 
 | | U11-48K CHHC Zn-finger protein domain | | Descriptor: | U11/U12 SMALL NUCLEAR RIBONUCLEOPROTEIN 48 KDA PROTEIN, ZINC ION | | Authors: | Tidow, H, Andreeva, A, Rutherford, T.J, Fersht, A.R. | | Deposit date: | 2008-07-18 | | Release date: | 2009-02-17 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the U11-48K CHHC zinc-finger domain that specifically binds the 5' splice site of U12-type introns.
Structure, 17, 2009
|
|
2WXC
 
 | | The folding mechanism of BBL: Plasticity of transition-state structure observed within an ultrafast folding protein family. | | Descriptor: | DIHYDROLIPOYLTRANSSUCCINASE | | Authors: | Neuweiler, H, Sharpe, T.D, Rutherford, T.J, Johnson, C.M, Allen, M.D, Ferguson, N, Fersht, A.R. | | Deposit date: | 2009-11-06 | | Release date: | 2009-11-17 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The Folding Mechanism of Bbl: Plasticity of Transition-State Structure Observed within an Ultrafast Folding Protein Family.
J.Mol.Biol., 390, 2009
|
|
1W0B
 
 | | Solution structure of the human alpha-hemoglobin stabilizing protein (AHSP) P30A mutant | | Descriptor: | ALPHA-HEMOGLOBIN STABILIZING PROTEIN | | Authors: | Santiveri, C.M, Perez-Canadillas, J.M, Vadivelu, M.K, Allen, M.D, Rutherford, T.J, Watkins, N.A, Bycroft, M. | | Deposit date: | 2004-06-01 | | Release date: | 2004-06-10 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the alpha-hemoglobin stabilizing protein: insights into conformational heterogeneity and binding.
J. Biol. Chem., 279, 2004
|
|
1W0A
 
 | | Solution structure of the trans form of the human alpha-hemoglobin stabilizing protein (AHSP) | | Descriptor: | ALPHA-HEMOGLOBIN STABILIZING PROTEIN | | Authors: | Santiveri, C.M, Perez-Canadillas, J.M, Vadivelu, M.K, Allen, M.D, Rutherford, T.J, Watkins, N.A, Bycroft, M. | | Deposit date: | 2004-06-01 | | Release date: | 2004-06-10 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the alpha-hemoglobin stabilizing protein: insights into conformational heterogeneity and binding.
J. Biol. Chem., 279, 2004
|
|
1W09
 
 | | Solution structure of the cis form of the human alpha-hemoglobin stabilizing protein (AHSP) | | Descriptor: | ALPHA-HEMOGLOBIN STABILIZING PROTEIN | | Authors: | Santiveri, C.M, Perez-Canadillas, J.M, Vadivelu, M.K, Allen, M.D, Rutherford, T.J, Watkins, N.A, Bycroft, M. | | Deposit date: | 2004-05-25 | | Release date: | 2004-06-10 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the alpha-hemoglobin stabilizing protein: insights into conformational heterogeneity and binding.
J. Biol. Chem., 279, 2004
|
|
6T8G
 
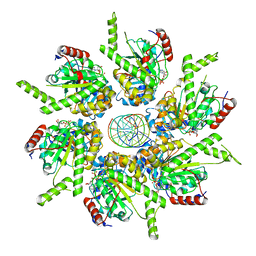 | | Stalled FtsK motor domain bound to dsDNA | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA translocase FtsK, dsDNA substrate | | Authors: | Jean, N.L, Lowe, J. | | Deposit date: | 2019-10-24 | | Release date: | 2019-11-20 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (4.34 Å) | | Cite: | FtsK in motion reveals its mechanism for double-stranded DNA translocation.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6T8B
 
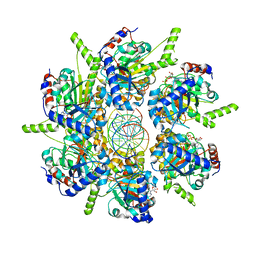 | | FtsK motor domain with dsDNA, translocating state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA translocase FtsK, MAGNESIUM ION, ... | | Authors: | Jean, N.L, Lowe, J. | | Deposit date: | 2019-10-24 | | Release date: | 2019-11-20 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.65 Å) | | Cite: | FtsK in motion reveals its mechanism for double-stranded DNA translocation.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6T8O
 
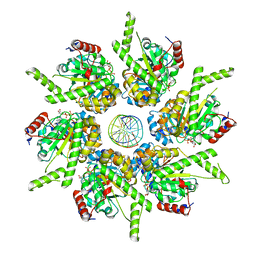 | | Stalled FtsK motor domain bound to dsDNA end | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA translocase FtsK, dsDNA substrate | | Authors: | Jean, N.L, Lowe, J. | | Deposit date: | 2019-10-24 | | Release date: | 2019-11-20 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.99 Å) | | Cite: | FtsK in motion reveals its mechanism for double-stranded DNA translocation.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7OQV
 
 | |
5LNP
 
 | |
7QCE
 
 | | Crystal structure of an atypical PHD finger of VIN3 | | Descriptor: | DI(HYDROXYETHYL)ETHER, VIN3 (Protein VERNALIZATION INSENSITIVE 3), ZINC ION | | Authors: | Franco-Echevarria, E, Fiedler, M, Dean, C, Bienz, M. | | Deposit date: | 2021-11-23 | | Release date: | 2022-11-16 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Plant vernalization proteins contain unusual PHD superdomains without histone H3 binding activity.
J.Biol.Chem., 298, 2022
|
|
7O06
 
 | |
7O0S
 
 | |
7O3B
 
 | |
7O6T
 
 | | Crystal structure of the polymerising VEL domain of VIN3 (R556D I575D mutant) | | Descriptor: | MAGNESIUM ION, Protein VERNALIZATION INSENSITIVE 3 | | Authors: | Fiedler, M, Franco-Echevarria, E, Dean, C, Bienz, M. | | Deposit date: | 2021-04-12 | | Release date: | 2022-11-09 | | Last modified: | 2022-11-23 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Head-to-tail polymerization by VEL proteins underpins cold-induced Polycomb silencing in flowering control.
Cell Rep, 41, 2022
|
|
7O6U
 
 | |
