1F2E
 
 | | STRUCTURE OF SPHINGOMONAD, GLUTATHIONE S-TRANSFERASE COMPLEXED WITH GLUTATHIONE | | Descriptor: | GLUTATHIONE, GLUTATHIONE S-TRANSFERASE | | Authors: | Nishio, T, Watanabe, T, Patel, A, Wang, Y, Lau, P.C.K, Grochulski, P, Li, Y, Cygler, M. | | Deposit date: | 2000-05-24 | | Release date: | 2000-06-21 | | Last modified: | 2011-12-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Properties of a Sphingomonad and Marine Bacterium Beta-Class Glutathione S-Transferases and Crystal Structure of the Former Complex with Glutathione
To be published
|
|
2ZK7
 
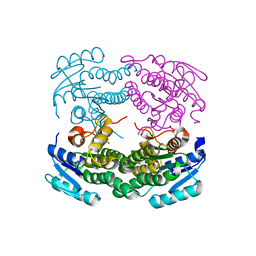 | | Structure of a C-terminal deletion mutant of Thermoplasma acidophilum aldohexose dehydrogenase (AldT) | | Descriptor: | Glucose 1-dehydrogenase related protein | | Authors: | Nishioka, T, Yasutake, Y, Nishiya, Y, Tamura, N, Tamura, T. | | Deposit date: | 2008-03-12 | | Release date: | 2009-01-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | C-terminal tail derived from the neighboring subunit is critical for the activity of Thermoplasma acidophilum D-aldohexose dehydrogenase
Proteins, 74, 2009
|
|
3AUS
 
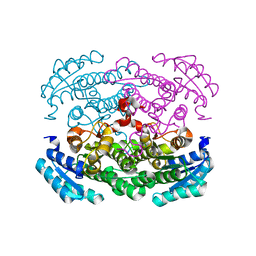 | |
3AUT
 
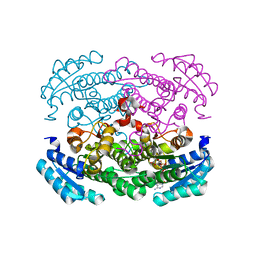 | | Crystal structure of Bacillus megaterium glucose dehydrogenase 4 in complex with NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Glucose 1-dehydrogenase 4 | | Authors: | Nishioka, T, Yasutake, Y, Nishiya, Y, Tamura, T. | | Deposit date: | 2011-02-16 | | Release date: | 2012-02-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-guided mutagenesis for the improvement of substrate specificity of Bacillus megaterium glucose 1-dehydrogenase IV
Febs J., 279, 2012
|
|
3AY6
 
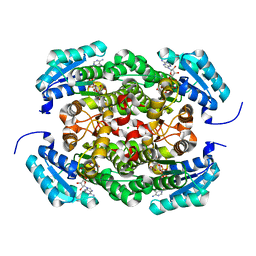 | | Crystal structure of Bacillus megaterium glucose dehydrogenase 4 A258F mutant in complex with NADH and D-glucose | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, CHLORIDE ION, Glucose 1-dehydrogenase 4, ... | | Authors: | Nishioka, T, Yasutake, Y, Nishiya, Y, Tamura, T. | | Deposit date: | 2011-04-29 | | Release date: | 2012-05-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-guided mutagenesis for the improvement of substrate specificity of Bacillus megaterium glucose 1-dehydrogenase IV
Febs J., 279, 2012
|
|
3AY7
 
 | | Crystal structure of Bacillus megaterium glucose dehydrogenase 4 G259A mutant | | Descriptor: | CHLORIDE ION, Glucose 1-dehydrogenase 4 | | Authors: | Nishioka, T, Yasutake, Y, Nishiya, Y, Tamura, T. | | Deposit date: | 2011-04-29 | | Release date: | 2012-05-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-guided mutagenesis for the improvement of substrate specificity of Bacillus megaterium glucose 1-dehydrogenase IV
Febs J., 279, 2012
|
|
3AUU
 
 | | Crystal structure of Bacillus megaterium glucose dehydrogenase 4 in complex with D-glucose | | Descriptor: | Glucose 1-dehydrogenase 4, beta-D-glucopyranose | | Authors: | Nishioka, T, Yasutake, Y, Nishiya, Y, Tamura, T. | | Deposit date: | 2011-02-16 | | Release date: | 2012-02-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-guided mutagenesis for the improvement of substrate specificity of Bacillus megaterium glucose 1-dehydrogenase IV
Febs J., 279, 2012
|
|
3VRM
 
 | | Structure of cytochrome P450 Vdh mutant T107A with bound vitamin D3 | | Descriptor: | (1S,3Z)-3-[(2E)-2-[(1R,3AR,7AS)-7A-METHYL-1-[(2R)-6-METHYLHEPTAN-2-YL]-2,3,3A,5,6,7-HEXAHYDRO-1H-INDEN-4-YLIDENE]ETHYLI DENE]-4-METHYLIDENE-CYCLOHEXAN-1-OL, PROTOPORPHYRIN IX CONTAINING FE, Vitamin D(3) 25-hydroxylase | | Authors: | Nishioka, T, Yasutake, Y, Tamura, T. | | Deposit date: | 2012-04-12 | | Release date: | 2013-04-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | A single mutation at the ferredoxin binding site of p450 vdh enables efficient biocatalytic production of 25-hydroxyvitamin d3.
Chembiochem, 14, 2013
|
|
2ZBO
 
 | | Crystal structure of low-redox-potential cytochrom c6 from brown alga Hizikia fusiformis at 1.6 A resolution | | Descriptor: | Cytochrome c6, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Akazaki, H, Kawai, F, Chida, H, Matsumoto, Y, Sirasaki, I, Nakade, H, Hirayama, M, Hosikawa, K, Suruga, K, Satoh, T, Yamada, S, Unzai, S, Hakamata, W, Nishio, T, Park, S.-Y, Oku, T. | | Deposit date: | 2007-10-26 | | Release date: | 2008-09-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Cloning, expression and purification of cytochrome c(6) from the brown alga Hizikia fusiformis and complete X-ray diffraction analysis of the structure
ACTA CRYSTALLOGR.,SECT.F, 64, 2008
|
|
7EOV
 
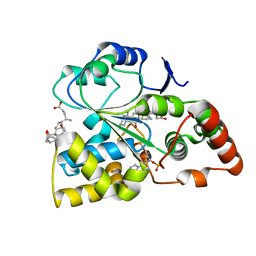 | | Crystal structure of mouse cytosolic sulfotransferase mSULT2A8 in complex with PAP and cholic acid | | Descriptor: | ADENOSINE-3'-5'-DIPHOSPHATE, CHOLIC ACID, cytosolic sulfotransferase SULT2A8 | | Authors: | Teramoto, T, Nishio, T, Kakuta, Y. | | Deposit date: | 2021-04-22 | | Release date: | 2021-05-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The crystal structure of mouse SULT2A8 reveals the mechanism of 7 alpha-hydroxyl, bile acid sulfation.
Biochem.Biophys.Res.Commun., 562, 2021
|
|
2DGE
 
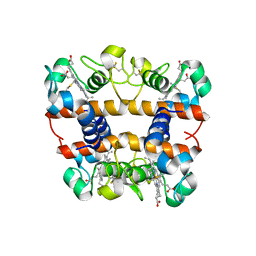 | | Crystal structure of oxidized cytochrome C6A from Arabidopsis thaliana | | Descriptor: | Cytochrome c6, PROTOPORPHYRIN IX CONTAINING FE, ZINC ION | | Authors: | Chida, H, Yokoyama, T, Kawai, F, Nakazawa, A, Akazaki, H, Takayama, Y, Hirano, T, Suruga, K, Satoh, T, Yamada, S, Kawachi, R, Unzai, S, Nishio, T, Park, S.-Y, Oku, T. | | Deposit date: | 2006-03-11 | | Release date: | 2006-07-04 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of oxidized cytochrome c(6A) from Arabidopsis thaliana
Febs Lett., 580, 2006
|
|
3DMI
 
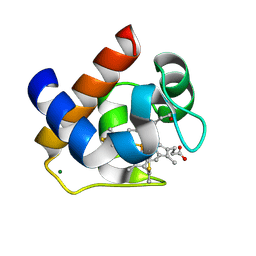 | | Crystallization and Structural Analysis of Cytochrome c6 from the Diatom Phaeodactylum tricornutum at 1.5 A resolution | | Descriptor: | HEME C, MAGNESIUM ION, cytochrome c6 | | Authors: | Akazaki, H, Kawai, F, Hosokawa, M, Hama, T, Hirano, T, Lim, B.-K, Sakurai, N, Hakamata, W, Park, S.-Y, Nishio, T, Oku, T. | | Deposit date: | 2008-07-01 | | Release date: | 2009-03-17 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystallization and structural analysis of cytochrome c(6) from the diatom Phaeodactylum tricornutum at 1.5 A resolution.
Biosci.Biotechnol.Biochem., 73, 2009
|
|
2ZZS
 
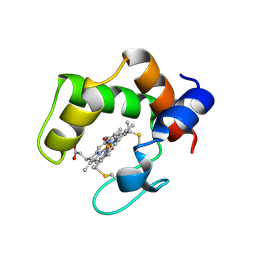 | | Crystal structure of cytochrome c554 from Vibrio parahaemolyticus strain RIMD2210633 | | Descriptor: | Cytochrome c554, GLYCEROL, HEME C | | Authors: | Akazaki, H, Kawai, F, Kumaki, Y, Sekine, K, Hakamata, W, Nishio, T, Park, S.-Y, Oku, T. | | Deposit date: | 2009-02-24 | | Release date: | 2010-03-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of cytochrome c554 from Vibrio parahaemolyticus strain RIMD2210633
To be Published
|
|
3WX7
 
 | | Crystal structure of COD | | Descriptor: | ACETATE ION, CALCIUM ION, Chitin oligosaccharide deacetylase, ... | | Authors: | Park, S.-Y, Sugiyama, K, Hirano, T, Nishio, T. | | Deposit date: | 2014-07-18 | | Release date: | 2014-12-03 | | Last modified: | 2019-04-03 | | Method: | X-RAY DIFFRACTION (1.349 Å) | | Cite: | Structure-based analysis of domain function of chitin oligosaccharide deacetylase from Vibrio parahaemolyticus.
FEBS Lett., 589, 2015
|
|
1GSH
 
 | | STRUCTURE OF ESCHERICHIA COLI GLUTATHIONE SYNTHETASE AT PH 7.5 | | Descriptor: | GLUTATHIONE BIOSYNTHETIC LIGASE | | Authors: | Matsuda, K, Kato, H, Yamaguchi, H, Nishioka, T, Katsube, Y, Oda, J. | | Deposit date: | 1995-05-16 | | Release date: | 1996-07-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of glutathione synthetase at optimal pH: domain architecture and structural similarity with other proteins.
Protein Eng., 9, 1996
|
|
1GSA
 
 | | STRUCTURE OF GLUTATHIONE SYNTHETASE COMPLEXED WITH ADP AND GLUTATHIONE | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GLUTATHIONE, GLUTATHIONE SYNTHETASE, ... | | Authors: | Hara, T, Kato, H, Nishioka, T, Katsube, Y, Oda, J. | | Deposit date: | 1995-06-08 | | Release date: | 1996-06-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A pseudo-michaelis quaternary complex in the reverse reaction of a ligase: structure of Escherichia coli B glutathione synthetase complexed with ADP, glutathione, and sulfate at 2.0 A resolution.
Biochemistry, 35, 1996
|
|
2GLT
 
 | | STRUCTURE OF ESCHERICHIA COLI GLUTATHIONE SYNTHETASE AT PH 6.0. | | Descriptor: | GLUTATHIONE BIOSYNTHETIC LIGASE | | Authors: | Matsuda, K, Yamaguchi, H, Kato, H, Nishioka, T, Katsube, Y, Oda, J. | | Deposit date: | 1995-05-16 | | Release date: | 1995-07-31 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of glutathione synthetase at optimal pH: domain architecture and structural similarity with other proteins.
Protein Eng., 9, 1996
|
|
3VTZ
 
 | |
1GDV
 
 | | CRYSTAL STRUCTURE OF CYTOCHROME C6 FROM RED ALGA PORPHYRA YEZOENSIS AT 1.57 A RESOLUTION | | Descriptor: | CYTOCHROME C6, HEME C | | Authors: | Yamada, S, Park, S.-Y, Shimizu, H, Shiro, Y, Oku, T. | | Deposit date: | 2000-10-06 | | Release date: | 2001-04-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Structure of cytochrome c6 from the red alga Porphyra yezoensis at 1. 57 A resolution.
Acta Crystallogr.,Sect.D, 56, 2000
|
|
11AS
 
 | |
1GLV
 
 | |
12AS
 
 | | ASPARAGINE SYNTHETASE MUTANT C51A, C315A COMPLEXED WITH L-ASPARAGINE AND AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, ASPARAGINE, ASPARAGINE SYNTHETASE | | Authors: | Nakatsu, T, Kato, H, Oda, J. | | Deposit date: | 1997-12-02 | | Release date: | 1998-12-30 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of asparagine synthetase reveals a close evolutionary relationship to class II aminoacyl-tRNA synthetase.
Nat.Struct.Biol., 5, 1998
|
|
3A4H
 
 | | Structure of cytochrome P450 vdh from Pseudonocardia autotrophica (orthorhombic crystal form) | | Descriptor: | CALCIUM ION, PROTOPORPHYRIN IX CONTAINING FE, Vitamin D hydroxylase | | Authors: | Yasutake, Y, Fujii, Y, Cheon, W.K, Arisawa, A, Tamura, T. | | Deposit date: | 2009-07-07 | | Release date: | 2010-07-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.06 Å) | | Cite: | Structural evidence for enhancement of sequential vitamin D3 hydroxylation activities by directed evolution of cytochrome P450 vitamin D3 hydroxylase
J.Biol.Chem., 285, 2010
|
|
3A51
 
 | | Structure of cytochrome P450 Vdh mutant (Vdh-K1) obtained by directed evolution with bound 25-hydroxyvitamin D3 | | Descriptor: | 3-{2-[1-(5-HYDROXY-1,5-DIMETHYL-HEXYL)-7A-METHYL-OCTAHYDRO-INDEN-4-YLIDENE]-ETHYLIDENE}-4-METHYLENE-CYCLOHEXANOL, ACETATE ION, CALCIUM ION, ... | | Authors: | Yasutake, Y, Fujii, Y, Cheon, W.K, Arisawa, A, Tamura, T. | | Deposit date: | 2009-07-24 | | Release date: | 2010-07-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural evidence for enhancement of sequential vitamin D3 hydroxylation activities by directed evolution of cytochrome P450 vitamin D3 hydroxylase
J.Biol.Chem., 285, 2010
|
|
3WEC
 
 | | Structure of P450 RauA (CYP1050A1) complexed with a biosynthetic intermediate of aurachin RE | | Descriptor: | 3-[(2E,6E,9R)-9-hydroxy-3,7,11-trimethyldodeca-2,6,10-trien-1-yl]-2-methylquinolin-4(1H)-one, Cytochrome P450, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Yasutake, Y, Kitagawa, W, Tamura, T. | | Deposit date: | 2013-07-03 | | Release date: | 2014-01-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Structure of the quinoline N-hydroxylating cytochrome P450 RauA, an essential enzyme that confers antibiotic activity on aurachin alkaloids
Febs Lett., 588, 2014
|
|
