2AE9
 
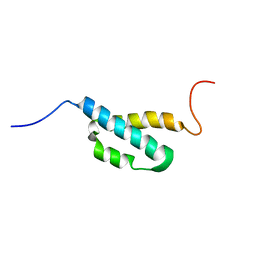 | | Solution Structure of the theta subunit of DNA polymerase III from E. coli | | Descriptor: | DNA polymerase III, theta subunit | | Authors: | Mueller, G.A, Kirby, T.W, Derose, E.F, Li, D, Schaaper, R.M, London, R.E. | | Deposit date: | 2005-07-21 | | Release date: | 2005-10-18 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Nuclear Magnetic Resonance Solution Structure of the Escherichia coli DNA Polymerase III {theta} Subunit.
J.Bacteriol., 187, 2005
|
|
3MQ1
 
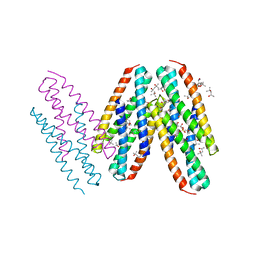 | | Crystal Structure of Dust Mite Allergen Der p 5 | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, Mite allergen Der p 5, ... | | Authors: | Mueller, G.A, Gosavi, R.A, Krahn, J.M, Edwards, L.L, Cuneo, M.J, Glesner, J, Pomes, A, Chapman, M.D, London, R.E, Pedersen, L.C. | | Deposit date: | 2010-04-27 | | Release date: | 2010-06-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Der p 5 crystal structure provides insight into the group 5 dust mite allergens.
J.Biol.Chem., 285, 2010
|
|
3OB4
 
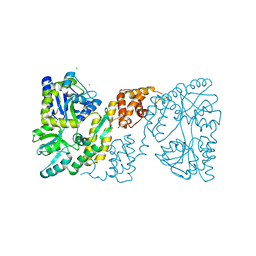 | | MBP-fusion protein of the major peanut allergen Ara h 2 | | Descriptor: | CHLORIDE ION, Maltose ABC transporter periplasmic protein,Arah 2, SULFATE ION, ... | | Authors: | Mueller, G.A, Gosavi, R.A, Moon, A.F, London, R.E, Pedersen, L.C. | | Deposit date: | 2010-08-06 | | Release date: | 2011-02-02 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.706 Å) | | Cite: | Ara h 2: crystal structure and IgE binding distinguish two subpopulations of peanut allergic patients by epitope diversity.
Allergy, 66, 2011
|
|
4OUO
 
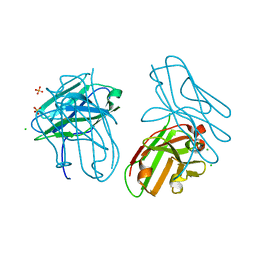 | | anti-Bla g 1 scFv | | Descriptor: | CHLORIDE ION, SULFATE ION, anti Bla g 1 scFv | | Authors: | Mueller, G.A, Ankney, J.A, Glesner, J, Khurana, T, Edwards, L.L, Pedersen, L.C, Perera, L, Slater, J.E, Pomes, A, London, R.E. | | Deposit date: | 2014-02-18 | | Release date: | 2014-03-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Characterization of an anti-Bla g 1 scFv: Epitope mapping and cross-reactivity.
Mol.Immunol., 59, 2014
|
|
4JRB
 
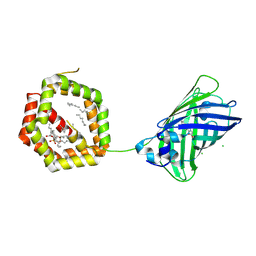 | | Structure of Cockroach Allergen Bla g 1 Tandem Repeat as a EGFP fusion | | Descriptor: | (1S)-2-{[{[(2R)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, CHLORIDE ION, DODECANE, ... | | Authors: | Mueller, G.A, Pedersen, L.C, Lih, F.B, Glesner, J, Moon, A.F, Chapman, M.D, Tomer, K, London, R.E. | | Deposit date: | 2013-03-21 | | Release date: | 2013-07-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.414 Å) | | Cite: | The novel structure of the cockroach allergen Bla g 1 has implications for allergenicity and exposure assessment.
J.Allergy Clin.Immunol., 132, 2013
|
|
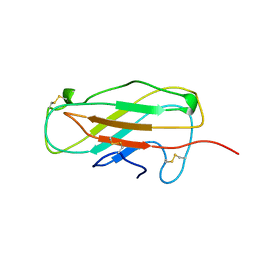
1A9V
 
 | |
2JW5
 
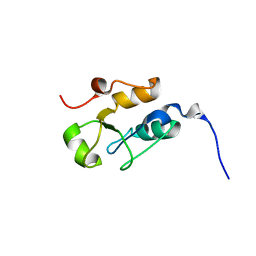 | |
2KHX
 
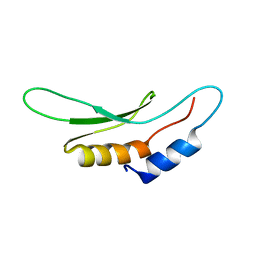 | | Drosha double-stranded RNA binding motif | | Descriptor: | Ribonuclease 3 | | Authors: | Mueller, G.A, Miller, M, Ghosh, M, DeRose, E.F, London, R.E, Hall, T. | | Deposit date: | 2009-04-13 | | Release date: | 2010-02-23 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the Drosha double-stranded RNA-binding domain.
Silence, 1, 2010
|
|
1EZP
 
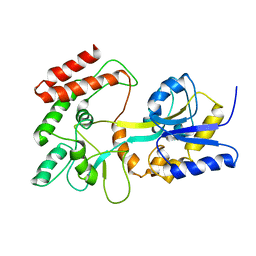 | | GLOBAL FOLD OF MALTODEXTRIN BINDING PROTEIN COMPLEXED WITH BETA-CYCLODEXTRIN USING PEPTIDE ORIENTATIONS FROM DIPOLAR COUPLINGS | | Descriptor: | MALTODEXTRIN BINDING PERIPLASMIC PROTEIN | | Authors: | Mueller, G.A, Choy, W.Y, Yang, D, Forman-Kay, J.D, Venters, R.A, Kay, L.E. | | Deposit date: | 2000-05-11 | | Release date: | 2001-05-08 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Global folds of proteins with low densities of NOEs using residual dipolar couplings: application to the 370-residue maltodextrin-binding protein.
J.Mol.Biol., 300, 2000
|
|
1EZO
 
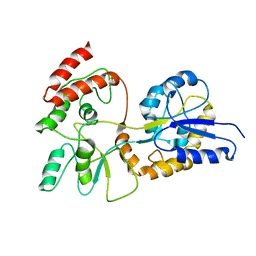 | | GLOBAL FOLD OF MALTODEXTRIN BINDING PROTEIN COMPLEXED WITH BETA-CYCLODEXTRIN | | Descriptor: | MALTOSE-BINDING PERIPLASMIC PROTEIN | | Authors: | Mueller, G.A, Choy, W.Y, Yang, D, Forman-Kay, J.D, Venters, R.A, Kay, L.E. | | Deposit date: | 2000-05-11 | | Release date: | 2001-05-03 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Global folds of proteins with low densities of NOEs using residual dipolar couplings: application to the 370-residue maltodextrin-binding protein.
J.Mol.Biol., 300, 2000
|
|
7LVE
 
 | |
7LVG
 
 | |
7LVF
 
 | |
7UV1
 
 | | Vicilin Ana o 1.0101 leader sequence residues 20-75 | | Descriptor: | Vicilin-like protein | | Authors: | Mueller, G.A, Foo, A.C.Y, DeRose, E.F. | | Deposit date: | 2022-04-29 | | Release date: | 2023-04-05 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Structure and IgE Cross-Reactivity among Cashew, Pistachio, Walnut, and Peanut Vicilin-Buried Peptides.
J.Agric.Food Chem., 71, 2023
|
|
7UV4
 
 | | Pis v 3.0101 vicilin leader sequence residues 56-115 | | Descriptor: | Vicilin Pis v 3.0101 | | Authors: | Mueller, G.A, Foo, A.C.Y, DeRose, E.F. | | Deposit date: | 2022-04-29 | | Release date: | 2023-04-05 | | Method: | SOLUTION NMR | | Cite: | Structure and IgE Cross-Reactivity among Cashew, Pistachio, Walnut, and Peanut Vicilin-Buried Peptides.
J.Agric.Food Chem., 71, 2023
|
|
7UV2
 
 | | Ana o 1 Leader Sequence Residues 82-132 | | Descriptor: | Vicilin-like protein | | Authors: | Mueller, G.A, Foo, A.C.Y, DeRose, E.F. | | Deposit date: | 2022-04-29 | | Release date: | 2023-04-05 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Structure and IgE Cross-Reactivity among Cashew, Pistachio, Walnut, and Peanut Vicilin-Buried Peptides.
J.Agric.Food Chem., 71, 2023
|
|
7UV3
 
 | | Pis v 3.0101 Vicilin Leader Sequence Residues 5-52 | | Descriptor: | Vicilin Pis v 3.0101 | | Authors: | Mueller, G.A, Foo, A.C.Y, DeRose, E.F. | | Deposit date: | 2022-04-29 | | Release date: | 2023-04-05 | | Method: | SOLUTION NMR | | Cite: | Structure and IgE Cross-Reactivity among Cashew, Pistachio, Walnut, and Peanut Vicilin-Buried Peptides.
J.Agric.Food Chem., 71, 2023
|
|
7V0V
 
 | | GFP Nanobody NMR Structure | | Descriptor: | Anti-GFP Nanobody | | Authors: | Mueller, G.A. | | Deposit date: | 2022-05-11 | | Release date: | 2022-06-08 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Nanobody Paratope Ensembles in Solution Characterized by MD Simulations and NMR.
Int J Mol Sci, 23, 2022
|
|
3H4Z
 
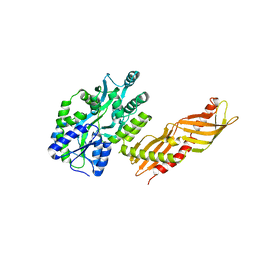 | | Crystal Structure of an MBP-Der p 7 fusion protein | | Descriptor: | Maltose-binding periplasmic protein fused with Allergen DERP7, SODIUM ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Pedersen, L.C, Mueller, G.A, London, R.E. | | Deposit date: | 2009-04-21 | | Release date: | 2010-03-31 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The structure of the dust mite allergen Der p 7 reveals similarities to innate immune proteins.
J.Allergy Clin.Immunol., 125, 2010
|
|
2MDP
 
 | |
4ZCE
 
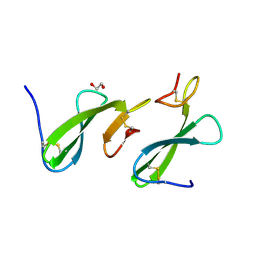 | | Crystal Structure of the dust mite allergen Der p 23 from Dermatophagoides pteronyssinus | | Descriptor: | 1,2-ETHANEDIOL, Dust mite allergen | | Authors: | Pedersen, L.C, Mueller, G.A, Randall, T.A, Glesner, J, Perera, L, Edwards, L.L, Chapman, M.D, London, R.E, Pomes, A. | | Deposit date: | 2015-04-15 | | Release date: | 2015-11-25 | | Last modified: | 2019-12-11 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Serological, genomic and structural analyses of the major mite allergen Der p 23.
Clin Exp Allergy, 46, 2016
|
|
8G4P
 
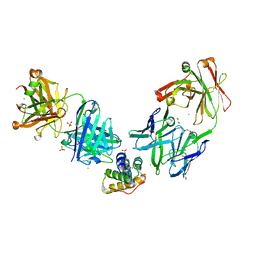 | | Crystal structure of the peanut allergen Ara h 2 bound by two neutralizing antibodies 13T1 and 13T5 | | Descriptor: | 1,2-ETHANEDIOL, 13T1 Fab light chain, 13T5 Fab heavy chain, ... | | Authors: | Pedersen, L.C, Mueller, G.A, Min, J. | | Deposit date: | 2023-02-10 | | Release date: | 2023-12-20 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Design of an Ara h 2 hypoallergen from conformational epitopes.
Clin Exp Allergy, 54, 2024
|
|
8SCF
 
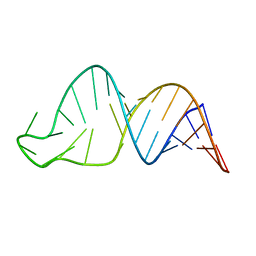 | | TCEIII NMR Structure | | Descriptor: | RNA (30-MER) | | Authors: | Warden, M.S, Mueller, G.A, Hall, T.M.T. | | Deposit date: | 2023-04-05 | | Release date: | 2023-07-12 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The translational repressor Glorund uses interchangeable RNA recognition domains to recognize Drosophila nanos.
Nucleic Acids Res., 51, 2023
|
|
8SCH
 
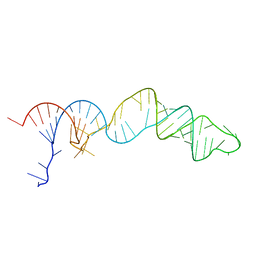 | | TCEI_III NMR Structure | | Descriptor: | RNA (68-MER) | | Authors: | Warden, M.S, Mueller, G.A, Hall, T.M.T. | | Deposit date: | 2023-04-05 | | Release date: | 2023-07-12 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR, SOLUTION SCATTERING | | Cite: | The translational repressor Glorund uses interchangeable RNA recognition domains to recognize Drosophila nanos.
Nucleic Acids Res., 51, 2023
|
|
4Q5R
 
 | |
