8SKW
 
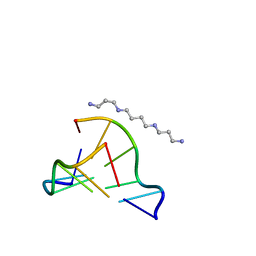 | | MicroED structure of d(CGCGCG)2 Z-DNA | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*CP*G)-3'), SPERMINE | | Authors: | Haymaker, A, Bardin, A.A, Martynowycz, M.W, Nannenga, B.L. | | Deposit date: | 2023-04-20 | | Release date: | 2023-08-16 | | Last modified: | 2023-12-20 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.1 Å) | | Cite: | Structure determination of a DNA crystal by MicroED.
Structure, 31, 2023
|
|
8SMK
 
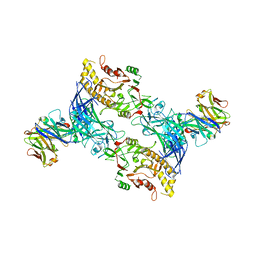 | | hPAD4 bound to Activating Fab hA362 | | Descriptor: | Activating Fab 362 heavy chain, Activating Fab 362 light chain, CALCIUM ION, ... | | Authors: | Maker, A, Verba, K.A. | | Deposit date: | 2023-04-26 | | Release date: | 2024-03-06 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Antibody discovery identifies regulatory mechanisms of protein arginine deiminase 4.
Nat.Chem.Biol., 20, 2024
|
|
8SML
 
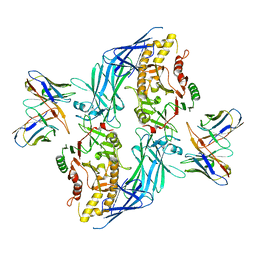 | | hPAD4 bound to inhibitory Fab hI365 | | Descriptor: | CALCIUM ION, Fab hI365 heavy chain, Fab hI365 light chain, ... | | Authors: | Maker, A, Verba, K.A. | | Deposit date: | 2023-04-26 | | Release date: | 2024-03-06 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Antibody discovery identifies regulatory mechanisms of protein arginine deiminase 4.
Nat.Chem.Biol., 20, 2024
|
|
7STZ
 
 | |
6VEL
 
 | |
6OZC
 
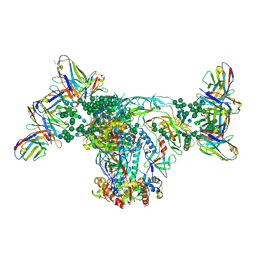 | | BG505 SOSIP.664 with 2G12 Fab2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2G12 Fab Heavy chain, ... | | Authors: | Cottrell, C.A, Ward, A.B. | | Deposit date: | 2019-05-15 | | Release date: | 2020-05-20 | | Last modified: | 2020-08-19 | | Method: | ELECTRON MICROSCOPY (3.79 Å) | | Cite: | Networks of HIV-1 Envelope Glycans Maintain Antibody Epitopes in the Face of Glycan Additions and Deletions.
Structure, 28, 2020
|
|
8VSU
 
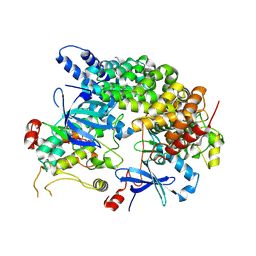 | | Cryo-EM structure of LKB1-STRADalpha-MO25alpha heterocomplex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Calcium-binding protein 39, Isoform 3 of STE20-related kinase adapter protein alpha, ... | | Authors: | Chan, L.M, Courteau, B.J, Verba, K.A. | | Deposit date: | 2024-01-24 | | Release date: | 2024-07-10 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.86 Å) | | Cite: | High-resolution single-particle imaging at 100-200 keV with the Gatan Alpine direct electron detector.
J.Struct.Biol., 216, 2024
|
|
5U03
 
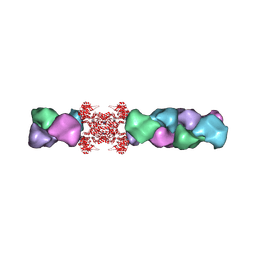 | | Cryo-EM structure of the human CTP synthase filament | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CTP synthase 1, URIDINE 5'-TRIPHOSPHATE | | Authors: | Lynch, E.M, Kollman, J.M. | | Deposit date: | 2016-11-22 | | Release date: | 2017-04-26 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (6.1 Å) | | Cite: | Human CTP synthase filament structure reveals the active enzyme conformation.
Nat. Struct. Mol. Biol., 24, 2017
|
|
5U05
 
 | |
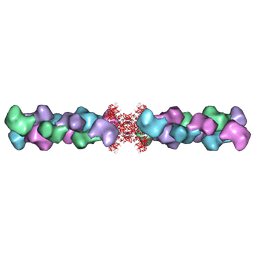
5U3C
 
 | |
5TKV
 
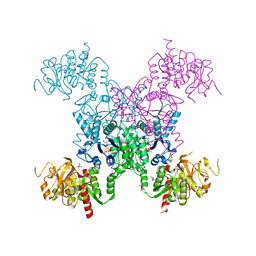 | | X-RAY CRYSTAL STRUCTURE OF THE "CLOSED" CONFORMATION OF CTP-INHIBITED E. COLI CYTIDINE TRIPHOSPHATE (CTP) SYNTHETASE | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, CTP synthase, ... | | Authors: | Baldwin, E.P, Endrizzi, J.A. | | Deposit date: | 2016-10-07 | | Release date: | 2017-04-26 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Human CTP synthase filament structure reveals the active enzyme conformation.
Nat. Struct. Mol. Biol., 24, 2017
|
|
5U6R
 
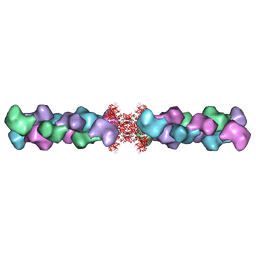 | |
2GHG
 
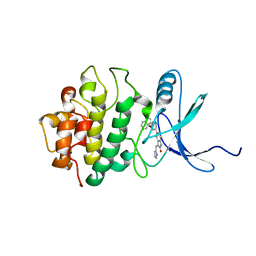 | | h-CHK1 complexed with A431994 | | Descriptor: | 5-{5-[(S)-2-AMINO-3-(1H-INDOL-3-YL)-PROPOXYL]-PYRIDIN-3-YL}-3-[1-(1H-PYRROL-2-YL)-METH-(Z)-YLIDENE]-1,3-DIHYDRO-INDOL-2-ONE, Serine/threonine-protein kinase Chk1 | | Authors: | Park, C. | | Deposit date: | 2006-03-27 | | Release date: | 2007-03-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Discovery and SAR of oxindole-pyridine-based protein kinase B/Akt inhibitors for treating cancers.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
