1UW1
 
 | |
3P5C
 
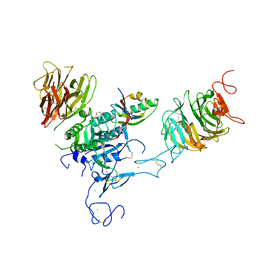 | | The structure of the LDLR/PCSK9 complex reveals the receptor in an extended conformation | | Descriptor: | CALCIUM ION, Low density lipoprotein receptor variant, Proprotein convertase subtilisin/kexin type 9 | | Authors: | Lo Surdo, P, Bottomley, M.J, Calzetta, A, Settembre, E.C, Cirillo, A, Pandit, S, Ni, Y, Hubbard, B, Sitlani, A, Carfi, A. | | Deposit date: | 2010-10-08 | | Release date: | 2011-10-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (4.2 Å) | | Cite: | Mechanistic implications for LDL receptor degradation from the PCSK9/LDLR structure at neutral pH.
Embo Rep., 12, 2011
|
|
3P5B
 
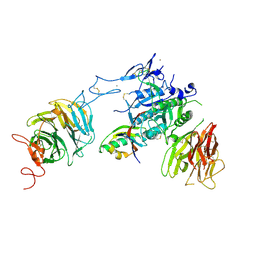 | | The structure of the LDLR/PCSK9 complex reveals the receptor in an extended conformation | | Descriptor: | CALCIUM ION, Low density lipoprotein receptor variant, Proprotein convertase subtilisin/kexin type 9 | | Authors: | Lo Surdo, P, Bottomley, M.J, Calzetta, A, Settembre, E.C, Cirillo, A, Pandit, S, Ni, Y, Hubbard, B, Sitlani, A, Carfi, A. | | Deposit date: | 2010-10-08 | | Release date: | 2011-10-26 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Mechanistic implications for LDL receptor degradation from the PCSK9/LDLR structure at neutral pH.
Embo Rep., 12, 2011
|
|
2W2M
 
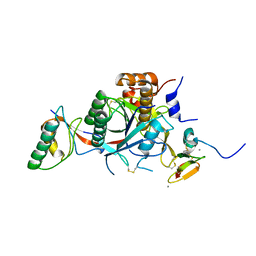 | | WT PCSK9-DELTAC BOUND TO WT EGF-A OF LDLR | | Descriptor: | CALCIUM ION, LOW-DENSITY LIPOPROTEIN RECEPTOR, PROPROTEIN CONVERTASE SUBTILISIN/KEXIN TYPE 9 | | Authors: | Bottomley, M.J, Cirillo, A, Orsatti, L, Ruggeri, L, Fisher, T.S, Santoro, J.C, Cummings, R.T, Cubbon, R.M, Lo Surdo, P, Calzetta, A, Noto, A, Baysarowich, J, Mattu, M, Talamo, F, De Francesco, R, Sparrow, C.P, Sitlani, A, Carfi, A. | | Deposit date: | 2008-11-03 | | Release date: | 2008-11-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and Biochemical Characterization of the Wild Type Pcsk9/Egf-Ab Complex and Natural Fh Mutants.
J.Biol.Chem., 284, 2009
|
|
2W2N
 
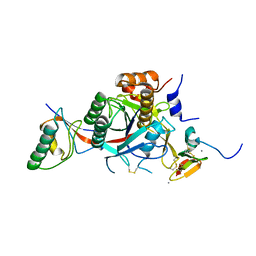 | | WT PCSK9-deltaC bound to EGF-A H306Y mutant of LDLR | | Descriptor: | CALCIUM ION, LOW-DENSITY LIPOPROTEIN RECEPTOR, PROPROTEIN CONVERTASE SUBTILISIN/KEXIN TYPE 9 | | Authors: | Bottomley, M.J, Cirillo, A, Orsatti, L, Ruggeri, L, Fisher, T.S, Santoro, J.C, Cummings, R.T, Cubbon, R.M, Lo Surdo, P, Calzetta, A, Noto, A, Baysarowich, J, Mattu, M, Talamo, F, De Francesco, R, Sparrow, C.P, Sitlani, A, Carfi, A. | | Deposit date: | 2008-11-03 | | Release date: | 2008-11-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and Biochemical Characterization of the Wild Type Pcsk9/Egf-Ab Complex and Natural Fh Mutants.
J.Biol.Chem., 284, 2009
|
|
2VQO
 
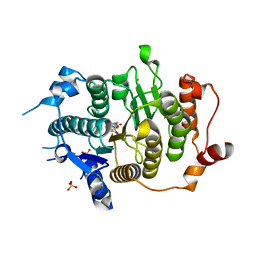 | | Structure of HDAC4 catalytic domain with a gain-of-function muation bound to a trifluoromethylketone inhbitor | | Descriptor: | 2,2,2-TRIFLUORO-1-{5-[(3-PHENYL-5,6-DIHYDROIMIDAZO[1,2-A]PYRAZIN-7(8H)-YL)CARBONYL]THIOPHEN-2-YL}ETHANE-1,1-DIOL, HISTONE DEACETYLASE 4, POTASSIUM ION, ... | | Authors: | Bottomley, M.J, Lo Surdo, P, Di Giovine, P, Cirillo, A, Scarpelli, R, Ferrigno, F, Jones, P, Neddermann, P, De Francesco, R, Steinkuhler, C, Gallinari, P, Carfi, A. | | Deposit date: | 2008-03-18 | | Release date: | 2008-07-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural and Functional Analysis of the Human Hdac4 Catalytic Domain Reveals a Regulatory Zinc-Binding Domain.
J.Biol.Chem., 283, 2008
|
|
2VQQ
 
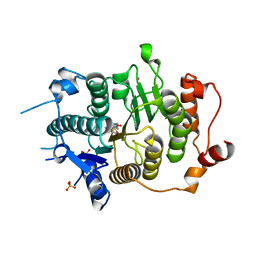 | | Structure of HDAC4 catalytic domain (a double cysteine-to-alanine mutant) bound to a trifluoromethylketone inhbitor | | Descriptor: | 2,2,2-TRIFLUORO-1-{5-[(3-PHENYL-5,6-DIHYDROIMIDAZO[1,2-A]PYRAZIN-7(8H)-YL)CARBONYL]THIOPHEN-2-YL}ETHANE-1,1-DIOL, HISTONE DEACETYLASE 4, POTASSIUM ION, ... | | Authors: | Bottomley, M.J, Lo Surdo, P, Di Giovine, P, Cirillo, A, Scarpelli, R, Ferrigno, F, Jones, P, Neddermann, P, De Francesco, R, Steinkuhler, C, Gallinari, P, Carfi, A. | | Deposit date: | 2008-03-18 | | Release date: | 2008-07-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and Functional Analysis of the Human Hdac4 Catalytic Domain Reveals a Regulatory Zinc-Binding Domain.
J.Biol.Chem., 283, 2008
|
|
2VQV
 
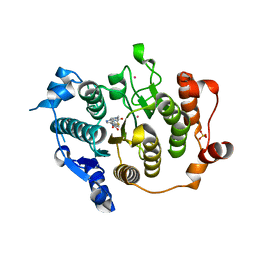 | | Structure of HDAC4 catalytic domain with a gain-of-function mutation bound to a hydroxamic acid inhibitor | | Descriptor: | HISTONE DEACETYLASE 4, N-hydroxy-5-[(3-phenyl-5,6-dihydroimidazo[1,2-a]pyrazin-7(8H)-yl)carbonyl]thiophene-2-carboxamide, POTASSIUM ION, ... | | Authors: | Bottomley, M.J, Lo Surdo, P, Di Giovine, P, Cirillo, A, Scarpelli, R, Ferrigno, F, Jones, P, Neddermann, P, De Francesco, R, Steinkuhler, C, Gallinari, P, Carfi, A. | | Deposit date: | 2008-03-19 | | Release date: | 2008-07-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural and Functional Analysis of the Human Hdac4 Catalytic Domain Reveals a Regulatory Structural Zinc-Binding Domain.
J.Biol.Chem., 283, 2008
|
|
2VQJ
 
 | | Structure of HDAC4 catalytic domain bound to a trifluoromethylketone inhbitor | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, 2,2,2-TRIFLUORO-1-{5-[(3-PHENYL-5,6-DIHYDROIMIDAZO[1,2-A]PYRAZIN-7(8H)-YL)CARBONYL]THIOPHEN-2-YL}ETHANE-1,1-DIOL, HISTONE DEACETYLASE 4, ... | | Authors: | Bottomley, M.J, Lo Surdo, P, Di Giovine, P, Cirillo, A, Scarpelli, R, Ferrigno, F, Jones, P, Neddermann, P, De Francesco, R, Steinkuhler, C, Gallinari, P, Carfi, A. | | Deposit date: | 2008-03-17 | | Release date: | 2008-07-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and Functional Analysis of the Human Hdac4 Catalytic Domain Reveals a Regulatory Zinc-Binding Domain.
J.Biol.Chem., 283, 2008
|
|
2VQW
 
 | | Structure of inhibitor-free HDAC4 catalytic domain (with gain-of- function mutation His332Tyr) | | Descriptor: | HISTONE DEACETYLASE 4, POTASSIUM ION, ZINC ION | | Authors: | Bottomley, M.J, Lo Surdo, P, Di Giovine, P, Cirillo, A, Scarpelli, R, Ferrigno, F, Jones, P, Neddermann, P, De Francesco, R, Steinkuhler, C, Gallinari, P, Carfi, A. | | Deposit date: | 2008-03-19 | | Release date: | 2008-08-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural and Functional Analysis of the Human Hdac4 Catalytic Domain Reveals a Regulatory Structural Zinc-Binding Domain.
J.Biol.Chem., 283, 2008
|
|
2VQM
 
 | | Structure of HDAC4 catalytic domain bound to a hydroxamic acid inhbitor | | Descriptor: | HISTONE DEACETYLASE 4, N-hydroxy-5-[(3-phenyl-5,6-dihydroimidazo[1,2-a]pyrazin-7(8H)-yl)carbonyl]thiophene-2-carboxamide, POTASSIUM ION, ... | | Authors: | Bottomley, M.J, Lo Surdo, P, Di Giovine, P, Cirillo, A, Scarpelli, R, Ferrigno, F, Jones, P, Neddermann, P, De Francesco, R, Steinkuhler, C, Gallinari, P, Carfi, A. | | Deposit date: | 2008-03-17 | | Release date: | 2008-07-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and Functional Analysis of the Human Hdac4 Catalytic Domain Reveals a Regulatory Zinc-Binding Domain.
J.Biol.Chem., 283, 2008
|
|
2W2Q
 
 | | PCSK9-deltaC D374H mutant bound to WT EGF-A of LDLR | | Descriptor: | CALCIUM ION, LOW-DENSITY LIPOPROTEIN RECEPTOR, PROPROTEIN CONVERTASE SUBTILISIN/KEXIN TYPE 9 | | Authors: | Bottomley, M.J, Cirillo, A, Orsatti, L, Ruggeri, L, Fisher, T.S, Santoro, J.C, Cummings, R.T, Cubbon, R.M, Lo Surdo, P, Calzetta, A, Noto, A, Baysarowich, J, Mattu, M, Talamo, F, De Francesco, R, Sparrow, C.P, Sitlani, A, Carfi, A. | | Deposit date: | 2008-11-03 | | Release date: | 2008-11-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Structural and Biochemical Characterization of the Wild Type Pcsk9/Egf-Ab Complex and Natural Fh Mutants.
J.Biol.Chem., 284, 2009
|
|
2W2P
 
 | | PCSK9-deltaC D374A mutant bound to WT EGF-A of LDLR | | Descriptor: | CALCIUM ION, LOW-DENSITY LIPOPROTEIN RECEPTOR, PROPROTEIN CONVERTASE SUBTILISIN/KEXIN TYPE 9 | | Authors: | Bottomley, M.J, Cirillo, A, Orsatti, L, Ruggeri, L, Fisher, T.S, Santoro, J.C, Cummings, R.T, Cubbon, R.M, Lo Surdo, P, Calzetta, A, Noto, A, Baysarowich, J, Mattu, M, Talamo, F, De Francesco, R, Sparrow, C.P, Sitlani, A, Carfi, A. | | Deposit date: | 2008-11-03 | | Release date: | 2008-11-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Structural and Biochemical Characterization of the Wild Type Pcsk9/Egf-Ab Complex and Natural Fh Mutants.
J.Biol.Chem., 284, 2009
|
|
2W2O
 
 | | PCSK9-deltaC D374Y mutant bound to WT EGF-A of LDLR | | Descriptor: | CALCIUM ION, LOW-DENSITY LIPOPROTEIN RECEPTOR, PROPROTEIN CONVERTASE SUBTILISIN/KEXIN TYPE 9 | | Authors: | Bottomley, M.J, Cirillo, A, Orsatti, L, Ruggeri, L, Fisher, T.S, Santoro, J.C, Cummings, R.T, Cubbon, R.M, Lo Surdo, P, Calzetta, A, Noto, A, Baysarowich, J, Mattu, M, Talamo, F, De Francesco, R, Sparrow, C.P, Sitlani, A, Carfi, A. | | Deposit date: | 2008-11-03 | | Release date: | 2008-11-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Structural and Biochemical Characterization of the Wild Type Pcsk9/Egf-Ab Complex and Natural Fh Mutants.
J.Biol.Chem., 284, 2009
|
|
6EUN
 
 | | Crystal structure of Neisseria meningitidis vaccine antigen NadA variant 3 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, 1-ETHOXY-2-(2-ETHOXYETHOXY)ETHANE, ... | | Authors: | Dello Iacono, L, Liguori, A, Malito, E, Bottomley, M.J. | | Deposit date: | 2017-10-30 | | Release date: | 2018-10-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | NadA3 Structures Reveal Undecad Coiled Coils and LOX1 Binding Regions Competed by Meningococcus B Vaccine-Elicited Human Antibodies.
MBio, 9, 2018
|
|
6H2Y
 
 | | human Fab 1E6 bound to fHbp variant 3 from Neisseria meningitidis serogroup B | | Descriptor: | 1,2-ETHANEDIOL, 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Veggi, D, Bianchi, F, Cozzi, R, Malito, E, Bottomley, M.J. | | Deposit date: | 2018-07-17 | | Release date: | 2019-08-14 | | Last modified: | 2020-02-26 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Cocrystal structure of meningococcal factor H binding protein variant 3 reveals a new crossprotective epitope recognized by human mAb 1E6.
Faseb J., 33, 2019
|
|
1B55
 
 | | PH DOMAIN FROM BRUTON'S TYROSINE KINASE IN COMPLEX WITH INOSITOL 1,3,4,5-TETRAKISPHOSPHATE | | Descriptor: | INOSITOL-(1,3,4,5)-TETRAKISPHOSPHATE, TYROSINE-PROTEIN KINASE BTK, ZINC ION | | Authors: | Djinovic Carugo, K, Baraldi, E, Hyvoenen, M, Lo Surdo, P, Riley, A.M, Potter, B.V.L, O'Brien, R, Ladbury, J.E, Saraste, M. | | Deposit date: | 1999-01-12 | | Release date: | 1999-06-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the PH domain from Bruton's tyrosine kinase in complex with inositol 1,3,4,5-tetrakisphosphate.
Structure Fold.Des., 7, 1999
|
|
1BWN
 
 | | PH DOMAIN AND BTK MOTIF FROM BRUTON'S TYROSINE KINASE MUTANT E41K IN COMPLEX WITH INS(1,3,4,5)P4 | | Descriptor: | BRUTON'S TYROSINE KINASE, INOSITOL-(1,3,4,5)-TETRAKISPHOSPHATE, ZINC ION | | Authors: | Djinovic Carugo, K, Baraldi, E, Hyvoenen, M, Lo Surdo, P, Riley, A, Potter, B, Saraste, M. | | Deposit date: | 1998-09-25 | | Release date: | 1999-06-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of the PH domain from Bruton's tyrosine kinase in complex with inositol 1,3,4,5-tetrakisphosphate.
Structure Fold.Des., 7, 1999
|
|
7NRU
 
 | |
5T5F
 
 | |
6CUJ
 
 | |
6XZW
 
 | |
6EUP
 
 | | Crystal structure of Neisseria meningitidis NadA variant 3 double mutant A33I-I38L | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, Adhesin A, ... | | Authors: | Dello Iacono, L, Liguori, A, Malito, E, Bottomley, M.J. | | Deposit date: | 2017-10-31 | | Release date: | 2018-10-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | NadA3 Structures Reveal Undecad Coiled Coils and LOX1 Binding Regions Competed by Meningococcus B Vaccine-Elicited Human Antibodies.
MBio, 9, 2018
|
|
5FLY
 
 | | The FhuD protein from S.pseudintermedius | | Descriptor: | 2,5,8,11,14,17,20,23-OCTAOXAPENTACOSAN-25-OL, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Malito, E. | | Deposit date: | 2015-10-29 | | Release date: | 2015-12-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.598 Å) | | Cite: | Crystal Structure of Fhud at 1.6 Angstrom Resolution: A Ferrichrome-Binding Protein from the Animal and Human Pathogen Staphylococcus Pseudintermedius
Acta Crystallogr.,Sect.F, 72, 2016
|
|
4OXR
 
 | | Structure of Staphylococcus pseudintermedius metal-binding protein SitA in complex with Manganese | | Descriptor: | MANGANESE (II) ION, Manganese ABC transporter, periplasmic-binding protein SitA | | Authors: | Abate, F, Malito, E, Bottomley, M. | | Deposit date: | 2014-02-06 | | Release date: | 2014-10-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Apo, Zn2+-bound and Mn2+-bound structures reveal ligand-binding properties of SitA from the pathogen Staphylococcus pseudintermedius.
Biosci.Rep., 34, 2014
|
|
