4E92
 
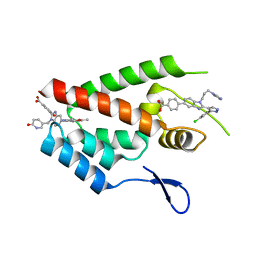 | | Crystal Structure of the N-Terminal Domain of HIV-1 Capsid in Complex With Inhibitor BM4 | | Descriptor: | 3-{5-[3-ethyl-5-(5-methylfuran-2-yl)-1H-pyrazol-1-yl]-1-[(6-oxo-1,6-dihydropyridin-3-yl)methyl]-1H-benzimidazol-2-yl}-4-hydroxybenzoic acid, 4-{2-[5-(3-chlorophenyl)-1H-pyrazol-4-yl]-1-[3-(1H-imidazol-1-yl)propyl]-1H-benzimidazol-5-yl}benzoic acid, Gag protein | | Authors: | Lemke, C.T. | | Deposit date: | 2012-03-20 | | Release date: | 2012-04-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Distinct Effects of Two HIV-1 Capsid Assembly Inhibitor Families That Bind the Same Site within the N-Terminal Domain of the Viral CA Protein.
J.Virol., 86, 2012
|
|
4E91
 
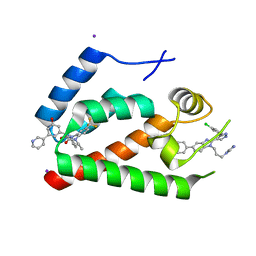 | | Crystal Structure of the N-Terminal Domain of HIV-1 Capsid in Complex With Inhibitor BD3 | | Descriptor: | (3S)-1-ethyl-3-[3-hydroxy-5-(pyridin-3-yl)phenyl]-5-phenyl-7-(trifluoromethyl)-1H-1,5-benzodiazepine-2,4(3H,5H)-dione, 4-{2-[5-(3-chlorophenyl)-1H-pyrazol-4-yl]-1-[3-(1H-imidazol-1-yl)propyl]-1H-benzimidazol-5-yl}benzoic acid, Gag protein, ... | | Authors: | Lemke, C.T. | | Deposit date: | 2012-03-20 | | Release date: | 2012-04-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Distinct Effects of Two HIV-1 Capsid Assembly Inhibitor Families That Bind the Same Site within the N-Terminal Domain of the Viral CA Protein.
J.Virol., 86, 2012
|
|
3P8N
 
 | | Crystal structure of HCV NS3/NS4A protease complexed with BI 201335 | | Descriptor: | HCV non-structural protein 4A, HCV serine protease NS3, N-[(cyclopentyloxy)carbonyl]-3-methyl-L-valyl-(4R)-4-[(8-bromo-7-methoxy-2-{2-[(2-methylpropanoyl)amino]-1,3-thiazol-4-yl}quinolin-4-yl)oxy]-N-[(1R,2S)-1-carboxy-2-ethenylcyclopropyl]-L-prolinamide, ... | | Authors: | Lemke, C.T. | | Deposit date: | 2010-10-14 | | Release date: | 2011-01-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Combined X-ray, NMR, and kinetic analyses reveal uncommon binding characteristics of the hepatitis C virus NS3-NS4A protease inhibitor BI 201335.
J.Biol.Chem., 286, 2011
|
|
3P8O
 
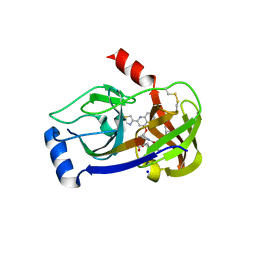 | | Crystal structure of HCV NS3/NS4A protease complexed with des-bromine analogue of BI 201335 | | Descriptor: | HCV non-structural protein 4A, HCV serine protease NS3, N-[(cyclopentyloxy)carbonyl]-3-methyl-L-valyl-(4R)-N-[(1R,2S)-1-carboxy-2-ethenylcyclopropyl]-4-[(7-methoxy-2-{2-[(2-methylpropanoyl)amino]-1,3-thiazol-4-yl}quinolin-4-yl)oxy]-L-prolinamide, ... | | Authors: | Lemke, C.T. | | Deposit date: | 2010-10-14 | | Release date: | 2011-01-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Combined X-ray, NMR, and kinetic analyses reveal uncommon binding characteristics of the hepatitis C virus NS3-NS4A protease inhibitor BI 201335.
J.Biol.Chem., 286, 2011
|
|
1KP2
 
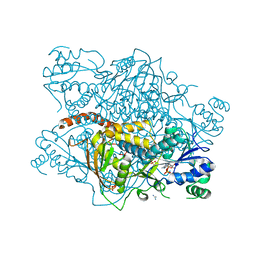 | |
1KP3
 
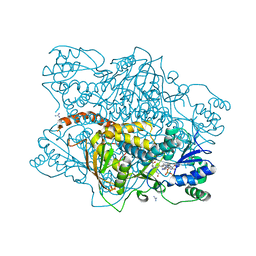 | |
1K97
 
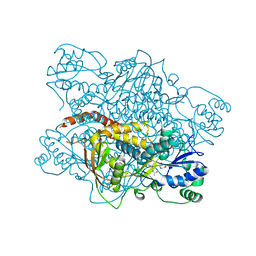 | |
1K92
 
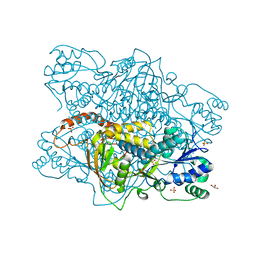 | |
4I32
 
 | | Crystal structure of HCV NS3/4A D168V protease complexed with compound 4 | | Descriptor: | (2R,6S,7E,10E,13aR,14aR,16aS)-2-{[7-methoxy-8-methyl-2-(propan-2-yloxy)quinolin-4-yl]oxy}-N-[(1-methylcyclopropyl)sulfonyl]-6-{[(1-methyl-1H-pyrazol-3-yl)carbonyl]amino}-5,16-dioxo-1,2,3,6,9,12,13,13a,14,15,16,16a-dodecahydrocyclopropa[e]pyrrolo[1,2-a][1,4]diazacyclopentadecine-14a(5H)-carboxamide, Genome polyprotein, HCV non-structural protein 4A, ... | | Authors: | Lemke, C.T. | | Deposit date: | 2012-11-23 | | Release date: | 2013-01-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.3001 Å) | | Cite: | Molecular Mechanism by Which a Potent Hepatitis C Virus NS3-NS4A Protease Inhibitor Overcomes Emergence of Resistance.
J.Biol.Chem., 288, 2013
|
|
4I33
 
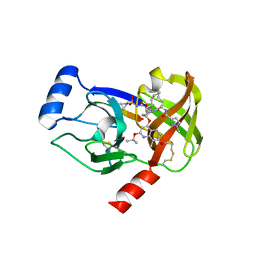 | | Crystal structure of HCV NS3/4A R155K protease complexed with compound 4 | | Descriptor: | (2R,6S,7E,10E,13aR,14aR,16aS)-2-{[7-methoxy-8-methyl-2-(propan-2-yloxy)quinolin-4-yl]oxy}-N-[(1-methylcyclopropyl)sulfonyl]-6-{[(1-methyl-1H-pyrazol-3-yl)carbonyl]amino}-5,16-dioxo-1,2,3,6,9,12,13,13a,14,15,16,16a-dodecahydrocyclopropa[e]pyrrolo[1,2-a][1,4]diazacyclopentadecine-14a(5H)-carboxamide, Genome polyprotein, HCV non-structural protein 4A, ... | | Authors: | Lemke, C.T. | | Deposit date: | 2012-11-23 | | Release date: | 2013-01-02 | | Last modified: | 2013-03-13 | | Method: | X-RAY DIFFRACTION (1.9001 Å) | | Cite: | Molecular Mechanism by Which a Potent Hepatitis C Virus NS3-NS4A Protease Inhibitor Overcomes Emergence of Resistance.
J.Biol.Chem., 288, 2013
|
|
4I31
 
 | | Crystal structure of HCV NS3/NS4A protease complexed with compound 4 | | Descriptor: | (2R,6S,7E,10E,13aR,14aR,16aS)-2-{[7-methoxy-8-methyl-2-(propan-2-yloxy)quinolin-4-yl]oxy}-N-[(1-methylcyclopropyl)sulfonyl]-6-{[(1-methyl-1H-pyrazol-3-yl)carbonyl]amino}-5,16-dioxo-1,2,3,6,9,12,13,13a,14,15,16,16a-dodecahydrocyclopropa[e]pyrrolo[1,2-a][1,4]diazacyclopentadecine-14a(5H)-carboxamide, Genome polyprotein, HCV non-structural protein 4A, ... | | Authors: | Lemke, C.T. | | Deposit date: | 2012-11-23 | | Release date: | 2013-01-02 | | Last modified: | 2013-03-13 | | Method: | X-RAY DIFFRACTION (1.9301 Å) | | Cite: | Molecular Mechanism by Which a Potent Hepatitis C Virus NS3-NS4A Protease Inhibitor Overcomes Emergence of Resistance.
J.Biol.Chem., 288, 2013
|
|
4J93
 
 | | Crystal Structure of the N-Terminal Domain of HIV-1 Capsid in Complex With Inhibitor BI-1 | | Descriptor: | (4S)-3-phenyl-4-(pyridin-3-yl)-4,5-dihydropyrrolo[3,4-c]pyrazol-6(2H)-one, 4-{2-[5-(3-chlorophenyl)-1H-pyrazol-4-yl]-1-[3-(1H-imidazol-1-yl)propyl]-1H-benzimidazol-5-yl}benzoic acid, Gag protein | | Authors: | Lemke, C.T. | | Deposit date: | 2013-02-15 | | Release date: | 2013-07-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Discovery of Novel Small-Molecule HIV-1 Replication Inhibitors That Stabilize Capsid Complexes.
Antimicrob.Agents Chemother., 57, 2013
|
|
4JMY
 
 | | Crystal structure of HCV NS3/NS4A protease complexed with DDIVPC peptide | | Descriptor: | HCV non-structural protein 4A, Polyprotein, SODIUM ION, ... | | Authors: | Lemke, C.T. | | Deposit date: | 2013-03-14 | | Release date: | 2013-10-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Importance of the peptide scaffold of drugs that target the hepatitis C virus NS3 protease and its crucial bioactive conformation and dynamic factors.
To be Published
|
|
8TXY
 
 | | X-ray crystal structure of JRD-SIK1/2i-3 bound to a MARK2-SIK2 chimera | | Descriptor: | DI(HYDROXYETHYL)ETHER, N-[(5P,8R)-5-(2-cyano-5-{[(3R)-1-methylpyrrolidin-3-yl]methoxy}pyridin-4-yl)pyrazolo[1,5-a]pyridin-2-yl]cyclopropanecarboxamide, SULFATE ION, ... | | Authors: | Raymond, D.D, Lemke, C.T, Shaffer, P.L, Collins, B, Steele, R, Seierstad, M. | | Deposit date: | 2023-08-24 | | Release date: | 2024-01-10 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Identification of highly selective SIK1/2 inhibitors that modulate innate immune activation and suppress intestinal inflammation.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8T6F
 
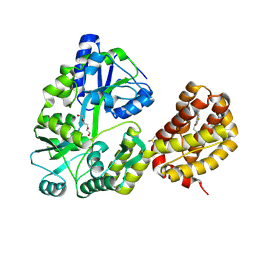 | | Crystal structure of human MBP-Myeloid cell leukemia 1 (Mcl-1) in complex with BRD810 inhibitor | | Descriptor: | (3aM,9S,15R)-4-chloro-3-ethyl-7-{3-[(6-fluoronaphthalen-1-yl)oxy]propyl}-2-methyl-15-[2-(morpholin-4-yl)ethyl]-2,10,11,12,13,15-hexahydropyrazolo[4',3':9,10][1,6]oxazacycloundecino[8,7,6-hi]indole-8-carboxylic acid, DIMETHYL SULFOXIDE, MAGNESIUM ION, ... | | Authors: | Poncet-Montange, G, Lemke, C.T. | | Deposit date: | 2023-06-15 | | Release date: | 2024-06-19 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | BRD-810 is a highly selective MCL1 inhibitor with optimized in vivo clearance and robust efficacy in solid and hematological tumor models.
Nat Cancer, 5, 2024
|
|
8AQN
 
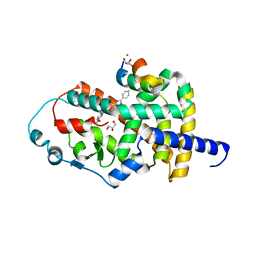 | | Crystal structure of PPARG and NCOR2 with BAY-4931, an inverse agonist (compound 6c) | | Descriptor: | 2-chloranyl-~{N}-[2-(4-ethylphenyl)-1,3-benzoxazol-5-yl]-5-nitro-benzamide, CALCIUM ION, GLYCEROL, ... | | Authors: | Friberg, A, Orsi, D.L, Pook, E, Braeuer, N, Lemke, C.T, Stellfeld, T, Puetter, V, Goldstein, J. | | Deposit date: | 2022-08-12 | | Release date: | 2022-11-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery and Structure-Based Design of Potent Covalent PPAR gamma Inverse-Agonists BAY-4931 and BAY-0069 .
J.Med.Chem., 65, 2022
|
|
8AQM
 
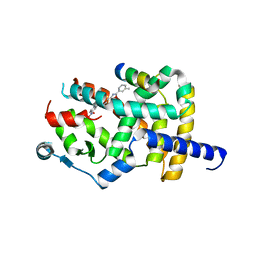 | | Crystal structure of PPARG and NCOR2 with an inverse agonist (compound 6a) | | Descriptor: | 2-chloranyl-~{N}-[2-(3-methylphenyl)-1,3-benzoxazol-5-yl]-5-nitro-benzamide, Nuclear receptor corepressor 2, Peroxisome proliferator-activated receptor gamma | | Authors: | Friberg, A, Orsi, D.L, Pook, E, Braeuer, N, Lemke, C.T, Stellfeld, T, Puetter, V, Goldstein, J. | | Deposit date: | 2022-08-12 | | Release date: | 2022-11-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery and Structure-Based Design of Potent Covalent PPAR gamma Inverse-Agonists BAY-4931 and BAY-0069 .
J.Med.Chem., 65, 2022
|
|
3I0O
 
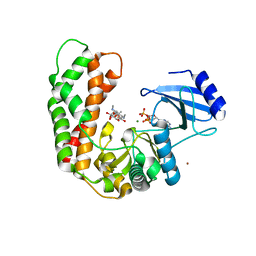 | | Crystal Structure of Spectinomycin Phosphotransferase, APH(9)-Ia, in complex with ADP and Spectinomcyin | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, NICKEL (II) ION, ... | | Authors: | Fong, D.H, Lemke, C.T, Hwang, J, Xiong, B, Berghuis, A.M. | | Deposit date: | 2009-06-25 | | Release date: | 2010-01-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the antibiotic resistance factor spectinomycin phosphotransferase from Legionella pneumophila.
J.Biol.Chem., 285, 2010
|
|
3I0Q
 
 | | Crystal Structure of the AMP-bound complex of Spectinomycin Phosphotransferase, APH(9)-Ia | | Descriptor: | ADENOSINE MONOPHOSPHATE, NICKEL (II) ION, Spectinomycin phosphotransferase | | Authors: | Berghuis, A.M, Fong, D.H, Lemke, C.T, Hwang, J.-Y, Xiong, B. | | Deposit date: | 2009-06-25 | | Release date: | 2010-01-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the antibiotic resistance factor spectinomycin phosphotransferase from Legionella pneumophila.
J.Biol.Chem., 285, 2010
|
|
3I1A
 
 | | Crystal Structure of apo Spectinomycin Phosphotransferase, APH(9)-Ia | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, DIMETHYL SULFOXIDE, Spectinomycin phosphotransferase, ... | | Authors: | Berghuis, A.M, Fong, D.H, Lemke, C.T, Hwang, J, Xiong, B. | | Deposit date: | 2009-06-25 | | Release date: | 2010-01-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of the antibiotic resistance factor spectinomycin phosphotransferase from Legionella pneumophila.
J.Biol.Chem., 285, 2010
|
|
8B94
 
 | | Crystal structure of PPARG and NCOR2 with BAY-5516, an inverse agonist | | Descriptor: | Nuclear receptor corepressor 2, Peroxisome proliferator-activated receptor gamma, ~{N}3-[4-[bis(fluoranyl)methoxy]-2-methyl-phenyl]-4-chloranyl-6-fluoranyl-~{N}1-[(4-fluorophenyl)methyl]benzene-1,3-dicarboxamide | | Authors: | Friberg, A, Orsi, D.L, Pook, E, Siegel, S, Lemke, C.T, Stellfeld, T, Puetter, V, Goldstein, J. | | Deposit date: | 2022-10-05 | | Release date: | 2022-12-28 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Discovery and characterization of orally bioavailable 4-chloro-6-fluoroisophthalamides as covalent PPARG inverse-agonists.
Bioorg.Med.Chem., 78, 2022
|
|
8B8X
 
 | | Crystal structure of PPARG and NCOR2 with SR10221, an inverse agonist | | Descriptor: | (2S)-2-{5-[(5-{[(1S)-1-(4-tert-butylphenyl)ethyl]carbamoyl}-2,3-dimethyl-1H-indol-1-yl)methyl]-2-chlorophenoxy}propanoic acid, Nuclear receptor corepressor 2, Peroxisome proliferator-activated receptor gamma | | Authors: | Friberg, A, Orsi, D.L, Pook, E, Siegel, S, Lemke, C.T, Stellfeld, T, Puetter, V, Goldstein, J. | | Deposit date: | 2022-10-05 | | Release date: | 2022-12-28 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Discovery and characterization of orally bioavailable 4-chloro-6-fluoroisophthalamides as covalent PPARG inverse-agonists.
Bioorg.Med.Chem., 78, 2022
|
|
8B92
 
 | | Crystal structure of PPARG and NCOR2 with an inverse agonist (compound SI-2) | | Descriptor: | 4-chloranyl-6-fluoranyl-~{N}3-[2-fluoranyl-4-(oxetan-3-yl)phenyl]-~{N}1-[(2-methoxyphenyl)methyl]benzene-1,3-dicarboxamide, Nuclear receptor corepressor 2, Peroxisome proliferator-activated receptor gamma, ... | | Authors: | Friberg, A, Orsi, D.L, Pook, E, Siegel, S, Lemke, C.T, Stellfeld, T, Puetter, V, Goldstein, J. | | Deposit date: | 2022-10-05 | | Release date: | 2022-12-28 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Discovery and characterization of orally bioavailable 4-chloro-6-fluoroisophthalamides as covalent PPARG inverse-agonists.
Bioorg.Med.Chem., 78, 2022
|
|
8B95
 
 | | Crystal structure of PPARG and NCOR2 with BAY-9683, an inverse agonist | | Descriptor: | Nuclear receptor corepressor 2, Peroxisome proliferator-activated receptor gamma, ~{N}1-[[3,4-bis(fluoranyl)phenyl]methyl]-4-chloranyl-6-fluoranyl-~{N}3-(3-methyl-5-morpholin-4-yl-pyridin-2-yl)benzene-1,3-dicarboxamide | | Authors: | Friberg, A, Orsi, D.L, Pook, E, Siegel, S, Lemke, C.T, Stellfeld, T, Puetter, V, Goldstein, J. | | Deposit date: | 2022-10-05 | | Release date: | 2022-12-28 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Discovery and characterization of orally bioavailable 4-chloro-6-fluoroisophthalamides as covalent PPARG inverse-agonists.
Bioorg.Med.Chem., 78, 2022
|
|
8B93
 
 | | Crystal structure of PPARG and NCOR2 with an inverse agonist (compound 15b) | | Descriptor: | 4-chloranyl-6-fluoranyl-~{N}1-[[4-fluoranyl-2-(2-methoxyethoxymethyl)phenyl]methyl]-~{N}3-[2-methyl-4-(trifluoromethyl)phenyl]benzene-1,3-dicarboxamide, Nuclear receptor corepressor 2, Peroxisome proliferator-activated receptor gamma | | Authors: | Friberg, A, Orsi, D.L, Pook, E, Siegel, S, Lemke, C.T, Stellfeld, T, Puetter, V, Goldstein, J. | | Deposit date: | 2022-10-05 | | Release date: | 2022-12-28 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Discovery and characterization of orally bioavailable 4-chloro-6-fluoroisophthalamides as covalent PPARG inverse-agonists.
Bioorg.Med.Chem., 78, 2022
|
|
