6E4D
 
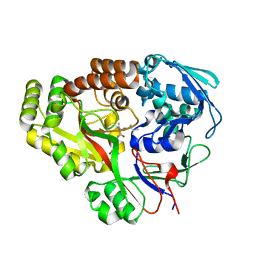 | | Atomic structure of Mycobacterium tuberculosis DppA | | Descriptor: | Periplasmic dipeptide-binding lipoprotein DPPA, VAL-VAL-VAL-ALA | | Authors: | Ko, Y, Mitra, A, Niederweis, M, Cingolani, G. | | Deposit date: | 2018-07-17 | | Release date: | 2019-09-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.252 Å) | | Cite: | Heme and hemoglobin utilization by Mycobacterium tuberculosis.
Nat Commun, 10, 2019
|
|
7JXT
 
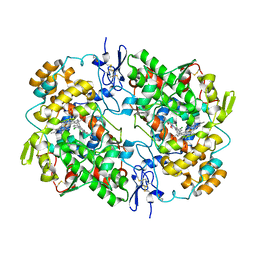 | | Ovine COX-1 in complex with the subtype-selective derivative 2a | | Descriptor: | 2-[4,5-bis(2-chlorophenyl)-1H-imidazol-2-yl]-6-(prop-2-en-1-yl)phenyl methoxyacetate, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Ko, Y, Iaselli, M, Miciaccia, M, Friedrich, L, Schneider, G, Scilimati, A, Cingolani, G. | | Deposit date: | 2020-08-27 | | Release date: | 2021-09-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Learning from Nature: From a Marine Natural Product to Synthetic Cyclooxygenase-1 Inhibitors by Automated De Novo Design.
Adv Sci, 8, 2021
|
|
6E3D
 
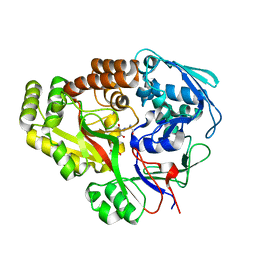 | | Atomic structure of Mycobacterium tuberculosis DppA | | Descriptor: | Periplasmic dipeptide-binding lipoprotein DPPA, tetra-peptide picked up from the expression host | | Authors: | Ko, Y, Mitra, A, Niederweis, M, Cingolani, G. | | Deposit date: | 2018-07-13 | | Release date: | 2019-09-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.271 Å) | | Cite: | Heme and hemoglobin utilization by Mycobacterium tuberculosis.
Nat Commun, 10, 2019
|
|
8GCN
 
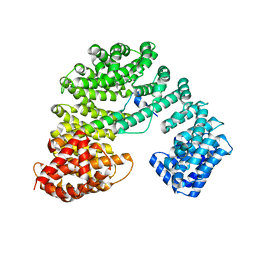 | |
6NKO
 
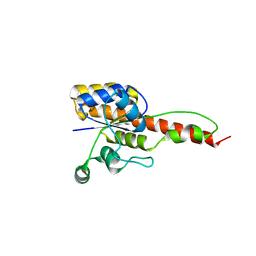 | | Crystal structure of ForH | | Descriptor: | ForH | | Authors: | Zheng, J, Irani, S, Zhang, Y. | | Deposit date: | 2019-01-07 | | Release date: | 2019-04-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.403 Å) | | Cite: | Identification of the Formycin A Biosynthetic Gene Cluster from Streptomyces kaniharaensis Illustrates the Interplay between Biological Pyrazolopyrimidine Formation and de Novo Purine Biosynthesis.
J. Am. Chem. Soc., 141, 2019
|
|
6VI2
 
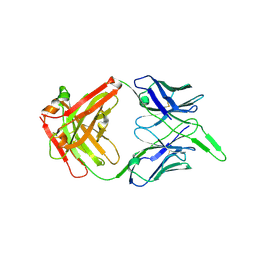 | | Structure of the unaligned Fab4 | | Descriptor: | FAB4 heavy chain, FAB4 light chain | | Authors: | Cingolani, G, Lokareddy, R, Ko, Y. | | Deposit date: | 2020-01-11 | | Release date: | 2020-09-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Recognition of an alpha-helical hairpin in P22 large terminase by a synthetic antibody fragment.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
6XMI
 
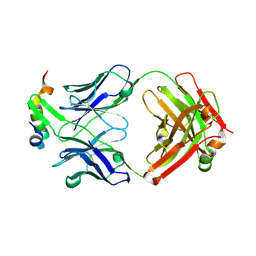 | | Structure of Fab4 bound to P22 TerL(1-33) | | Descriptor: | Fab Heavy chain, Fab Light chain, Terminase, ... | | Authors: | Cingolani, G, Lokareddy, R, Ko, Y. | | Deposit date: | 2020-06-30 | | Release date: | 2020-08-19 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Recognition of an alpha-helical hairpin in P22 large terminase by a synthetic antibody fragment.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
6VI1
 
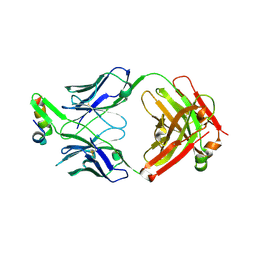 | | Structure of Fab4 bound to P22 TerL(1-33) | | Descriptor: | Synthetic Fab4 heavy chain, Synthetic Fab4 light chain, Terminase, ... | | Authors: | Cingolani, G, Lokareddy, R, Ko, Y. | | Deposit date: | 2020-01-11 | | Release date: | 2020-09-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Recognition of an alpha-helical hairpin in P22 large terminase by a synthetic antibody fragment.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
7WZX
 
 | | The structure of a Twitch Radical SAM Dehydrogenase SpeY | | Descriptor: | (2~{S},4~{S},6~{R})-2-[(2~{S},3~{R},5~{S},6~{R})-3,5-bis(methylamino)-2,4,6-tris(oxidanyl)cyclohexyl]oxy-6-methyl-4-oxidanyl-oxan-3-one, 4Fe-4S cluster-binding domain-containing protein, GLYCEROL, ... | | Authors: | Hou, X.L, Zhou, J.H. | | Deposit date: | 2022-02-19 | | Release date: | 2022-12-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.980013 Å) | | Cite: | Dioxane Bridge Formation during the Biosynthesis of Spectinomycin Involves a Twitch Radical S -Adenosyl Methionine Dehydrogenase That May Have Evolved from an Epimerase.
J.Am.Chem.Soc., 144, 2022
|
|
7X0B
 
 | | The structure of a Twitch Radical SAM Dehydrogenase SpeY | | Descriptor: | 4Fe-4S cluster-binding domain-containing protein, CHLORIDE ION, GLYCEROL, ... | | Authors: | Hou, X.L, Zhou, J.H. | | Deposit date: | 2022-02-21 | | Release date: | 2022-12-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.02027535 Å) | | Cite: | Dioxane Bridge Formation during the Biosynthesis of Spectinomycin Involves a Twitch Radical S -Adenosyl Methionine Dehydrogenase That May Have Evolved from an Epimerase.
J.Am.Chem.Soc., 144, 2022
|
|
7WZV
 
 | | The structure of a Twitch Radical SAM Dehydrogenase SpeY | | Descriptor: | (1~{S},2~{R},4~{S},5~{R})-2,4-bis(methylamino)-6-[(2~{S},3~{R},4~{S},6~{R})-6-methyl-3,4-bis(oxidanyl)oxan-2-yl]oxy-cyclohexane-1,3,5-triol, 1,2-ETHANEDIOL, 4Fe-4S cluster-binding domain-containing protein, ... | | Authors: | Zhou, J.H, Hou, X.L. | | Deposit date: | 2022-02-19 | | Release date: | 2022-12-28 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.899313 Å) | | Cite: | Dioxane Bridge Formation during the Biosynthesis of Spectinomycin Involves a Twitch Radical S -Adenosyl Methionine Dehydrogenase That May Have Evolved from an Epimerase.
J.Am.Chem.Soc., 144, 2022
|
|
