6GRL
 
 | |
6HZN
 
 | | Crystal structure of human dermatan sulfate epimerase 1 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Hasan, M, Unge, J, Westergren-Thorsson, G, Ellervik, U, Mueller, U, Malmstrom, A, Tykesson, E. | | Deposit date: | 2018-10-23 | | Release date: | 2020-01-22 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | The structure of human dermatan sulfate epimerase 1 emphasizes the importance of C5-epimerization of glucuronic acid in higher organisms
Chem Sci, 2020
|
|
5OLK
 
 | | Crystal structure of the ATP-cone-containing NrdB from Leeuwenhoekiella blandensis | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, MAGNESIUM ION, ... | | Authors: | Hasan, M, Grinberg, A.R, Sjoberg, B.M, Logan, D.T. | | Deposit date: | 2017-07-28 | | Release date: | 2018-01-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Novel ATP-cone-driven allosteric regulation of ribonucleotide reductase via the radical-generating subunit.
Elife, 7, 2018
|
|
6SF5
 
 | | Mn-containing form of the ribonucleotide reductase NrdB protein from Leeuwenhoekiella blandensis | | Descriptor: | MANGANESE (II) ION, Ribonucleoside-diphosphate reductase, beta subunit 1 | | Authors: | Hasan, M, Rozman Grinberg, I, Sjoberg, B.M, Logan, D.T. | | Deposit date: | 2019-08-01 | | Release date: | 2019-08-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Class Id ribonucleotide reductase utilizes a Mn2(IV,III) cofactor and undergoes large conformational changes on metal loading.
J.Biol.Inorg.Chem., 24, 2019
|
|
6SF4
 
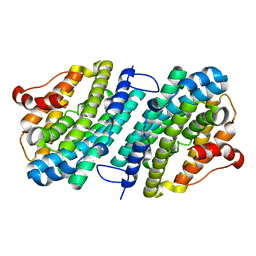 | | Apo form of the ribonucleotide reductase NrdB protein from Leeuwenhoekiella blandensis | | Descriptor: | Ribonucleoside-diphosphate reductase, beta subunit 1 | | Authors: | Hasan, M, Rozman Grinberg, I, Sjoberg, B.M, Logan, D.T. | | Deposit date: | 2019-07-31 | | Release date: | 2019-08-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Class Id ribonucleotide reductase utilizes a Mn2(IV,III) cofactor and undergoes large conformational changes on metal loading.
J.Biol.Inorg.Chem., 24, 2019
|
|
2XQ0
 
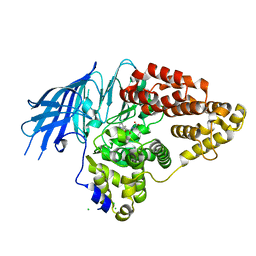 | | Structure of yeast LTA4 hydrolase in complex with Bestatin | | Descriptor: | 2-(3-AMINO-2-HYDROXY-4-PHENYL-BUTYRYLAMINO)-4-METHYL-PENTANOIC ACID, CHLORIDE ION, LEUKOTRIENE A-4 HYDROLASE, ... | | Authors: | Helgstrand, C, Hasan, M, Usyal, H, Haeggstrom, J.Z, Thunnissen, M.M.G.M. | | Deposit date: | 2010-08-31 | | Release date: | 2010-12-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.955 Å) | | Cite: | A Leukotriene A(4) Hydrolase-Related Aminopeptidase from Yeast Undergoes Induced Fit Upon Inhibitor Binding.
J.Mol.Biol., 406, 2011
|
|
2XPY
 
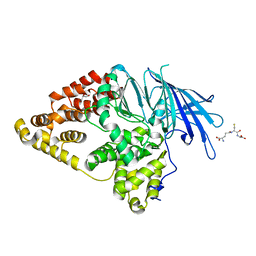 | | Structure of Native Leukotriene A4 Hydrolase from Saccharomyces cerevisiae | | Descriptor: | GLUTATHIONE, GLYCEROL, LEUKOTRIENE A-4 HYDROLASE, ... | | Authors: | Helgstrand, C, Hasan, M, Usyal, H, Haeggstrom, J.Z, Thunnissen, M.M.G.M. | | Deposit date: | 2010-08-31 | | Release date: | 2010-12-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | A Leukotriene A(4) Hydrolase-Related Aminopeptidase from Yeast Undergoes Induced Fit Upon Inhibitor Binding.
J.Mol.Biol., 406, 2011
|
|
2XPZ
 
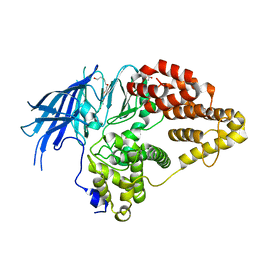 | | Structure of native yeast LTA4 hydrolase | | Descriptor: | (R,R)-2,3-BUTANEDIOL, LEUKOTRIENE A-4 HYDROLASE, POLYETHYLENE GLYCOL (N=34), ... | | Authors: | Helgstrand, C, Hasan, M, Usyal, H, Haeggstrom, J.Z, Thunnissen, M.M.G.M. | | Deposit date: | 2010-08-31 | | Release date: | 2010-12-29 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A Leukotriene A(4) Hydrolase-Related Aminopeptidase from Yeast Undergoes Induced Fit Upon Inhibitor Binding.
J.Mol.Biol., 406, 2011
|
|
7P97
 
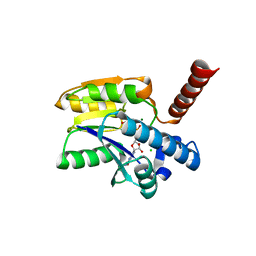 | | Structure of 3-phospho-D-glycerate guanylyltransferase with product 3-GPPG bound | | Descriptor: | 3-(guanosine-5'-diphospho)-D-glycerate, 3-phospho-D-glycerate guanylyltransferase, CHLORIDE ION, ... | | Authors: | Palm, G.J, Berndt, L, Lammers, M. | | Deposit date: | 2021-07-26 | | Release date: | 2022-02-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Diversification by CofC and Control by CofD Govern Biosynthesis and Evolution of Coenzyme F 420 and Its Derivative 3PG-F 420.
Mbio, 13, 2022
|
|
4GAA
 
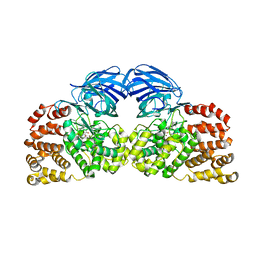 | | Structure of Leukotriene A4 hydrolase from Xenopus laevis complexed with inhibitor bestatin | | Descriptor: | 2-(3-AMINO-2-HYDROXY-4-PHENYL-BUTYRYLAMINO)-4-METHYL-PENTANOIC ACID, MGC78867 protein, ZINC ION | | Authors: | Stsiapanava, A, Kumar, R.B, Haeggstrom, J.Z, Rinaldo-Matthis, A. | | Deposit date: | 2012-07-25 | | Release date: | 2013-06-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Product formation controlled by substrate dynamics in leukotriene A4 hydrolase.
Biochim.Biophys.Acta, 1844, 2014
|
|
7SZ2
 
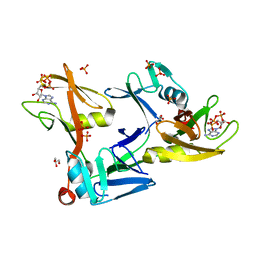 | | Mouse PARP13/ZAP ZnF5-WWE1-WWE2 bound to ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Ayanath Kuttiyatveetil, J.R, Pascal, J.M. | | Deposit date: | 2021-11-25 | | Release date: | 2022-10-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures and functional analysis of the ZnF5-WWE1-WWE2 region of PARP13/ZAP define a distinctive mode of engaging poly(ADP-ribose).
Cell Rep, 41, 2022
|
|
7SZ3
 
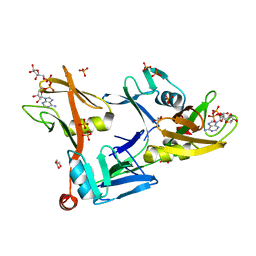 | | Mouse PARP13/ZAP ZnF5-WWE1-WWE2 bound to ADPr | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Ayanath Kuttiyatveetil, J.R, Pascal, J.M. | | Deposit date: | 2021-11-25 | | Release date: | 2022-10-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures and functional analysis of the ZnF5-WWE1-WWE2 region of PARP13/ZAP define a distinctive mode of engaging poly(ADP-ribose).
Cell Rep, 41, 2022
|
|
7Q6C
 
 | | complement C6 FIM1-2 bound to CP010 antibody | | Descriptor: | ACETATE ION, CP010 heavy chain, CP010 light chain, ... | | Authors: | Olesen, H.G, Andersen, G.R. | | Deposit date: | 2021-11-06 | | Release date: | 2022-05-25 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.29274 Å) | | Cite: | Development, Characterization, and in vivo Validation of a Humanized C6 Monoclonal Antibody that Inhibits the Membrane Attack Complex.
J Innate Immun, 2022
|
|
7Q6Y
 
 | | The X-ray crystal structure of CbTan2, a tannase enzyme from Clostridium butyricum | | Descriptor: | 1,2-ETHANEDIOL, Alpha/beta hydrolase, DI(HYDROXYETHYL)ETHER | | Authors: | Coleman, T, Mazurkewich, S, Larsbrink, J. | | Deposit date: | 2021-11-09 | | Release date: | 2022-03-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Structural diversity and substrate preferences of three tannase enzymes encoded by the anaerobic bacterium Clostridium butyricum.
J.Biol.Chem., 298, 2022
|
|
