2XKJ
 
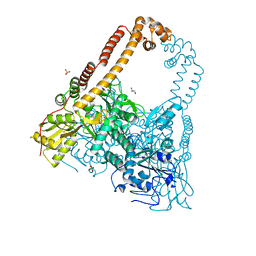 | | CRYSTAL STRUCTURE OF CATALYTIC CORE OF A. BAUMANNII TOPO IV (PARE- PARC FUSION TRUNCATE) | | Descriptor: | GLYCEROL, SULFATE ION, TOPOISOMERASE IV | | Authors: | Wohlkonig, A, Chan, P.F, Fosberry, A.P, Homes, P, Huang, J, Kranz, M, Leydon, V.R, Miles, T.J, Pearson, N.D, Perera, R.L, Shillings, A.J, Gwynn, M.N, Bax, B.D. | | Deposit date: | 2010-07-08 | | Release date: | 2010-09-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Basis of Quinolone Inhibition of Type Iia Topoisomerases and Target-Mediated Resistance
Nat.Struct.Mol.Biol., 17, 2010
|
|
2XKK
 
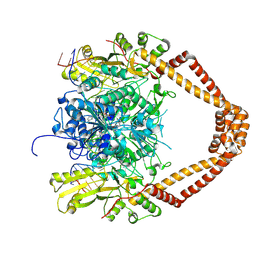 | | CRYSTAL STRUCTURE OF MOXIFLOXACIN, DNA, and A. BAUMANNII TOPO IV (PARE-PARC FUSION TRUNCATE) | | Descriptor: | 1-cyclopropyl-6-fluoro-8-methoxy-7-[(4aS,7aS)-octahydro-6H-pyrrolo[3,4-b]pyridin-6-yl]-4-oxo-1,4-dihydroquinoline-3-carboxylic acid, DNA, MAGNESIUM ION, ... | | Authors: | Wohlkonig, A, Chan, P.F, Fosberry, A.P, Homes, P, Huang, J, Kranz, M, Leydon, V.R, Miles, T.J, Pearson, N.D, Perera, R.L, Shillings, A.J, Gwynn, M.N, Bax, B.D. | | Deposit date: | 2010-07-08 | | Release date: | 2010-09-01 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Structural Basis of Quinolone Inhibition of Type Iia Topoisomerases and Target-Mediated Resistance
Nat.Struct.Mol.Biol., 17, 2010
|
|
5CDM
 
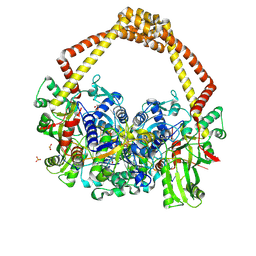 | | 2.5A structure of QPT-1 with S.aureus DNA gyrase and DNA | | Descriptor: | (2R,4S,4aS)-2,4-dimethyl-8-nitro-1,2,4,4a-tetrahydro-2'H,6H-spiro[1,4-oxazino[4,3-a]quinoline-5,5'-pyrimidine]-2',4',6'(1'H,3'H)-trione, DNA (5'-D(P*GP*AP*GP*CP*GP*TP*AP*C*GP*GP*CP*CP*GP*TP*AP*CP*GP*CP*TP*T)-3'), DNA gyrase subunit A, ... | | Authors: | Bax, B.D, Srikannathasan, V, Chan, P.F. | | Deposit date: | 2015-07-04 | | Release date: | 2015-12-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis of DNA gyrase inhibition by antibacterial QPT-1, anticancer drug etoposide and moxifloxacin.
Nat Commun, 6, 2015
|
|
5CDN
 
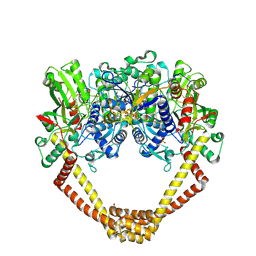 | | 2.8A structure of etoposide with S.aureus DNA gyrase and DNA | | Descriptor: | (5S,5aR,8aR,9R)-9-(4-hydroxy-3,5-dimethoxyphenyl)-8-oxo-5,5a,6,8,8a,9-hexahydrofuro[3',4':6,7]naphtho[2,3-d][1,3]dioxol -5-yl 4,6-O-[(1R)-ethylidene]-beta-D-glucopyranoside, DNA (5'-D(P*GP*AP*GP*CP*GP*TP*AP**GP*GP*CP*CP*GP*TP*AP*CP*GP*CP*TP*C)-3'), DNA (5'-D(P*GP*AP*GP*CP*GP*TP*AP*C*GP*GP*CP*CP*GP*TP*AP*CP*GP*CP*TP*C)-3'), ... | | Authors: | Bax, B.D, Srikannathasan, V, Chan, P.F. | | Deposit date: | 2015-07-04 | | Release date: | 2015-12-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Structural basis of DNA gyrase inhibition by antibacterial QPT-1, anticancer drug etoposide and moxifloxacin.
Nat Commun, 6, 2015
|
|
5CDQ
 
 | | 2.95A structure of Moxifloxacin with S.aureus DNA gyrase and DNA | | Descriptor: | 1-cyclopropyl-6-fluoro-8-methoxy-7-[(4aS,7aS)-octahydro-6H-pyrrolo[3,4-b]pyridin-6-yl]-4-oxo-1,4-dihydroquinoline-3-carboxylic acid, DNA (5'-D(P*GP*AP*GP*CP*GP*TP*AP*T*GP*GP*CP*CP*AP*TP*AP*CP*GP*CP*TP*T)-3'), DNA gyrase subunit A, ... | | Authors: | Bax, B.D, Srikannathasan, V, Chan, P.F. | | Deposit date: | 2015-07-04 | | Release date: | 2015-12-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structural basis of DNA gyrase inhibition by antibacterial QPT-1, anticancer drug etoposide and moxifloxacin.
Nat Commun, 6, 2015
|
|
5CDO
 
 | | 3.15A structure of QPT-1 with S.aureus DNA gyrase and DNA | | Descriptor: | (2R,4S,4aS)-4',6'-dihydroxy-2,4-dimethyl-8-nitro-1,2,4,4a-tetrahydro-2'H,6H-spiro[1,4-oxazino[4,3-a]quinoline-5,5'-pyrimidin]-2'-one, (2R,4S,4aS,5R)-6'-hydroxy-2,4-dimethyl-8-nitro-1,2,4,4a-tetrahydro-2'H,6H-spiro[1,4-oxazino[4,3-a]quinoline-5,5'-pyrimidine]-2',4'(3'H)-dione, (2R,4S,4aS,5S)-6'-hydroxy-2,4-dimethyl-8-nitro-1,2,4,4a-tetrahydro-2'H,6H-spiro[1,4-oxazino[4,3-a]quinoline-5,5'-pyrimidine]-2',4'(3'H)-dione, ... | | Authors: | Bax, B.D, Srikannathasan, V, Chan, P.F. | | Deposit date: | 2015-07-04 | | Release date: | 2015-12-16 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Structural basis of DNA gyrase inhibition by antibacterial QPT-1, anticancer drug etoposide and moxifloxacin.
Nat Commun, 6, 2015
|
|
5CDR
 
 | | 2.65 structure of S.aureus DNA gyrase and artificially nicked DNA | | Descriptor: | DNA (5'-D(*AP*GP*CP*CP*GP*TP*AP*)-3'), DNA (5'-D(*AP*GP*CP*CP*GP*TP*AP*GP*GP*TP*AP*CP*CP*TP*AP*CP*GP*GP*CP*T)-3'), DNA (5'-D(*GP*GP*TP*AP*CP*CP*TP*AP*CP*GP*GP*CP*T)-3'), ... | | Authors: | Bax, B.D, Srikannathasan, V, Chan, P.F. | | Deposit date: | 2015-07-04 | | Release date: | 2015-12-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural basis of DNA gyrase inhibition by antibacterial QPT-1, anticancer drug etoposide and moxifloxacin.
Nat Commun, 6, 2015
|
|
5CDP
 
 | | 2.45A structure of etoposide with S.aureus DNA gyrase and DNA | | Descriptor: | (5S,5aR,8aR,9R)-9-(4-hydroxy-3,5-dimethoxyphenyl)-8-oxo-5,5a,6,8,8a,9-hexahydrofuro[3',4':6,7]naphtho[2,3-d][1,3]dioxol -5-yl 4,6-O-[(1R)-ethylidene]-beta-D-glucopyranoside, DNA (5'-D(*AP*GP*CP*CP*GP*TP*AP*G*GP*GP*TP*AP*CP*CP*TP*AP*CP*GP*GP*CP*T)-3'), DNA gyrase subunit A, ... | | Authors: | Bax, B.D, Srikannathasan, V, Chan, P.F. | | Deposit date: | 2015-07-04 | | Release date: | 2015-12-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural basis of DNA gyrase inhibition by antibacterial QPT-1, anticancer drug etoposide and moxifloxacin.
Nat Commun, 6, 2015
|
|
1I2Z
 
 | | E. COLI ENOYL REDUCTASE IN COMPLEX WITH NAD AND BRL-12654 | | Descriptor: | 4-(2-THIENYL)-1-(4-METHYLBENZYL)-1H-IMIDAZOLE, ENOYL-[ACYL-CARRIER-PROTEIN] REDUCTASE [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Heerding, D.A, Miller, W.H, Payne, D.J, Janson, C.A, Qiu, X. | | Deposit date: | 2001-02-12 | | Release date: | 2002-02-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | 1,4-Disubstituted imidazoles are potential antibacterial agents functioning as inhibitors of enoyl acyl carrier protein
reductase (FabI).
Bioorg.Med.Chem.Lett., 11, 2001
|
|
1I30
 
 | | E. Coli Enoyl Reductase +NAD+SB385826 | | Descriptor: | 1,3,4,9-TETRAHYDRO-2-(HYDROXYBENZOYL)-9-[(4-HYDROXYPHENYL)METHYL]-6-METHOXY-2H-PYRIDO[3,4-B]INDOLE, ENOYL-[ACYL-CARRIER-PROTEIN] REDUCTASE [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Seefeld, M.A, Miller, W.H, Payne, D.J, Janson, C.A, Qiu, X. | | Deposit date: | 2001-02-12 | | Release date: | 2002-02-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Inhibitors of bacterial enoyl acyl carrier protein reductase (FabI): 2,9-disubstituted 1,2,3,4-tetrahydropyrido[3,4-b]indoles as potential antibacterial agents.
Bioorg.Med.Chem.Lett., 11, 2001
|
|
