8T4S
 
 | |
8CWO
 
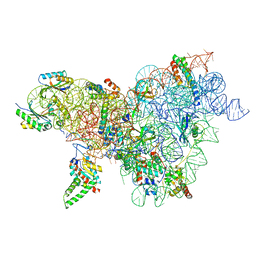 | | Cutibacterium acnes 30S ribosomal subunit with Sarecycline bound, body domain only in the local refined map | | Descriptor: | 16S ribosomal RNA, 30S ribosomal protein S11, 30S ribosomal protein S12, ... | | Authors: | Lomakin, I.B, Devarkar, S.C, Bunick, C.G. | | Deposit date: | 2022-05-19 | | Release date: | 2023-03-15 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.84 Å) | | Cite: | Sarecycline inhibits protein translation in Cutibacterium acnes 70S ribosome using a two-site mechanism.
Nucleic Acids Res., 51, 2023
|
|
8CVO
 
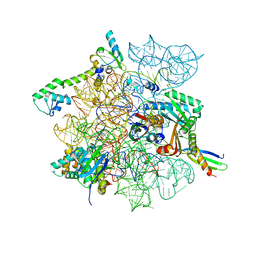 | | Cutibacterium acnes 30S ribosomal subunit with Sarecycline bound, head domain only in the local refined map | | Descriptor: | 16S ribosomal RNA, 30S ribosomal protein S10, 30S ribosomal protein S13, ... | | Authors: | Lomakin, I.B, Devarkar, S.C, Bunick, C.G. | | Deposit date: | 2022-05-18 | | Release date: | 2023-03-15 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | Sarecycline inhibits protein translation in Cutibacterium acnes 70S ribosome using a two-site mechanism.
Nucleic Acids Res., 51, 2023
|
|
8CVM
 
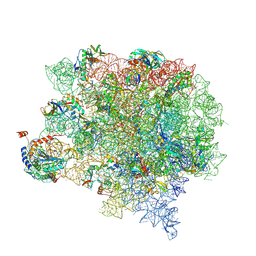 | | Cutibacterium acnes 50S ribosomal subunit with P-site tRNA and Sarecycline bound in the local refined map | | Descriptor: | 23S ribosomal RNA, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | Authors: | Lomakin, I.B, Devarkar, S.C, Bunick, C.G. | | Deposit date: | 2022-05-18 | | Release date: | 2023-03-15 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.66 Å) | | Cite: | Sarecycline inhibits protein translation in Cutibacterium acnes 70S ribosome using a two-site mechanism.
Nucleic Acids Res., 51, 2023
|
|
9C4G
 
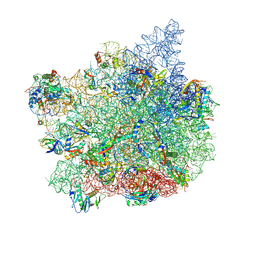 | | Cutibacterium acnes 50S ribosomal subunit with Clindamycin bound | | Descriptor: | 23S rRNA, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | Authors: | Lomakin, I.B, Devarkar, S.C, Bunick, C.G. | | Deposit date: | 2024-06-04 | | Release date: | 2024-08-21 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.53 Å) | | Cite: | Mechanistic Basis for the Translation Inhibition of Cutibacterium acnes by Clindamycin.
J Invest Dermatol., 144, 2024
|
|
8CRX
 
 | | Cutibacterium acnes 70S ribosome with mRNA, P-site tRNA and Sarecycline bound | | Descriptor: | 23S ribosomal RNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Lomakin, I.B, Devarkar, S.C, Bunick, C.G. | | Deposit date: | 2022-05-12 | | Release date: | 2023-03-22 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.78 Å) | | Cite: | Sarecycline inhibits protein translation in Cutibacterium acnes 70S ribosome using a two-site mechanism.
Nucleic Acids Res., 51, 2023
|
|
5F98
 
 | | Crystal structure of RIG-I in complex with Cap-0 RNA | | Descriptor: | 7N-METHYL-8-HYDROGUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Probable ATP-dependent RNA helicase DDX58, ... | | Authors: | Wang, C, Marcotrigiano, J, Miller, M, Jiang, F. | | Deposit date: | 2015-12-09 | | Release date: | 2016-01-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.28 Å) | | Cite: | Structural basis for m7G recognition and 2'-O-methyl discrimination in capped RNAs by the innate immune receptor RIG-I.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5F9F
 
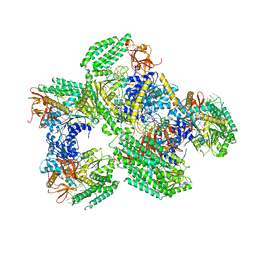 | | Crystal structure of RIG-I helicase-RD in complex with 24-mer blunt-end hairpin RNA | | Descriptor: | (R,R)-2,3-BUTANEDIOL, MAGNESIUM ION, Probable ATP-dependent RNA helicase DDX58, ... | | Authors: | Wang, C, Marcotrigiano, J, Miller, M.T, Jiang, F. | | Deposit date: | 2015-12-09 | | Release date: | 2016-01-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.601 Å) | | Cite: | Structural basis for m7G recognition and 2'-O-methyl discrimination in capped RNAs by the innate immune receptor RIG-I.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5F9H
 
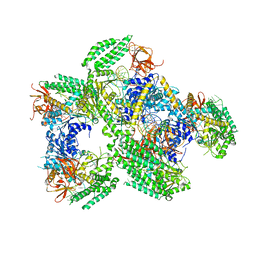 | | Crystal structure of RIG-I helicase-RD in complex with 24-mer 5' triphosphate hairpin RNA | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Probable ATP-dependent RNA helicase DDX58, ... | | Authors: | Wang, C, Marcotrigiano, J, Miller, M, Jiang, F. | | Deposit date: | 2015-12-09 | | Release date: | 2016-01-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural basis for m7G recognition and 2'-O-methyl discrimination in capped RNAs by the innate immune receptor RIG-I.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
9D6E
 
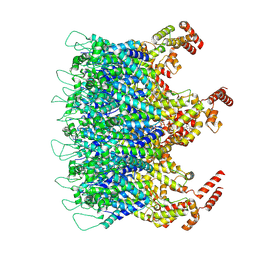 | | Gag CA-SP1 immature lattice bound with Bevirimat from enveloped virus like particles | | Descriptor: | 3alpha-[(3-carboxy-3-methylbutanoyl)oxy]-8alpha,9beta,10alpha,13alpha,17alpha,19beta-lup-20(29)-en-28-oic acid, Gag polyprotein, INOSITOL HEXAKISPHOSPHATE | | Authors: | Wu, C, Meuser, M.E, Xiong, Y. | | Deposit date: | 2024-08-14 | | Release date: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.09 Å) | | Cite: | Structural insights into inhibitor mechanisms on immature HIV-1 Gag lattice revealed by high-resolution in situ single-particle cryo-EM.
Biorxiv, 2024
|
|
9CWV
 
 | |
9D6D
 
 | | Gag CA-SP1 immature lattice bound with Lenacapavir from enveloped virus like particles | | Descriptor: | Gag polyprotein, INOSITOL HEXAKISPHOSPHATE, N-[(1S)-1-(3-{4-chloro-3-[(methylsulfonyl)amino]-1-(2,2,2-trifluoroethyl)-1H-indazol-7-yl}-6-[3-methyl-3-(methylsulfonyl)but-1-yn-1-yl]pyridin-2-yl)-2-(3,5-difluorophenyl)ethyl]-2-[(3bS,4aR)-5,5-difluoro-3-(trifluoromethyl)-3b,4,4a,5-tetrahydro-1H-cyclopropa[3,4]cyclopenta[1,2-c]pyrazol-1-yl]acetamide | | Authors: | Wu, C, Meuser, M.E, Xiong, Y. | | Deposit date: | 2024-08-14 | | Release date: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.18 Å) | | Cite: | Structural insights into inhibitor mechanisms on immature HIV-1 Gag lattice revealed by high-resolution in situ single-particle cryo-EM.
Biorxiv, 2024
|
|
9D88
 
 | |
9D6C
 
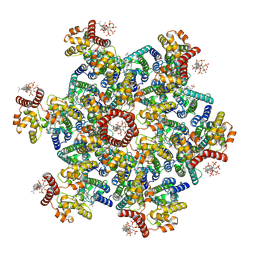 | | Gag CA-SP1 immature lattice bound with Lenacapavir and Bevirimat from enveloped virus like particles | | Descriptor: | 3alpha-[(3-carboxy-3-methylbutanoyl)oxy]-8alpha,9beta,10alpha,13alpha,17alpha,19beta-lup-20(29)-en-28-oic acid, Gag, INOSITOL HEXAKISPHOSPHATE, ... | | Authors: | Wu, C, Meuser, M.E, Xiong, Y. | | Deposit date: | 2024-08-14 | | Release date: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.1 Å) | | Cite: | Structural insights into inhibitor mechanisms on immature HIV-1 Gag lattice revealed by high-resolution in situ single-particle cryo-EM.
Biorxiv, 2024
|
|
9DWD
 
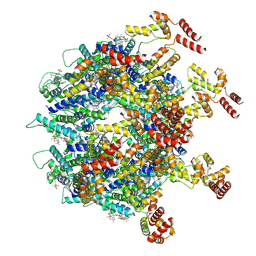 | | Gag CA-SP1 immature lattice bound with Lenacapavir from enveloped virus like particles (T8I) | | Descriptor: | Gag, INOSITOL HEXAKISPHOSPHATE, N-[(1S)-1-(3-{4-chloro-3-[(methylsulfonyl)amino]-1-(2,2,2-trifluoroethyl)-1H-indazol-7-yl}-6-[3-methyl-3-(methylsulfonyl)but-1-yn-1-yl]pyridin-2-yl)-2-(3,5-difluorophenyl)ethyl]-2-[(3bS,4aR)-5,5-difluoro-3-(trifluoromethyl)-3b,4,4a,5-tetrahydro-1H-cyclopropa[3,4]cyclopenta[1,2-c]pyrazol-1-yl]acetamide | | Authors: | Wu, C, Meuser, M.E, Xiong, Y. | | Deposit date: | 2024-10-09 | | Release date: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.13 Å) | | Cite: | Structural insights into inhibitor mechanisms on immature HIV-1 Gag lattice revealed by high-resolution in situ single-particle cryo-EM.
Biorxiv, 2024
|
|
7JQB
 
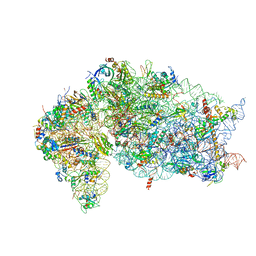 | | SARS-CoV-2 Nsp1 and rabbit 40S ribosome complex | | Descriptor: | 40S ribosomal protein S21, 40S ribosomal protein S24, 40S ribosomal protein S26, ... | | Authors: | Yuan, S, Xiong, Y. | | Deposit date: | 2020-08-10 | | Release date: | 2020-12-02 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Nonstructural Protein 1 of SARS-CoV-2 Is a Potent Pathogenicity Factor Redirecting Host Protein Synthesis Machinery toward Viral RNA.
Mol.Cell, 80, 2020
|
|
7JQC
 
 | | SARS-CoV-2 Nsp1, CrPV IRES and rabbit 40S ribosome complex | | Descriptor: | 40S ribosomal protein S21, 40S ribosomal protein S24, 40S ribosomal protein S26, ... | | Authors: | Yuan, S, Xiong, Y. | | Deposit date: | 2020-08-10 | | Release date: | 2020-12-02 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Nonstructural Protein 1 of SARS-CoV-2 Is a Potent Pathogenicity Factor Redirecting Host Protein Synthesis Machinery toward Viral RNA.
Mol.Cell, 80, 2020
|
|
7UIM
 
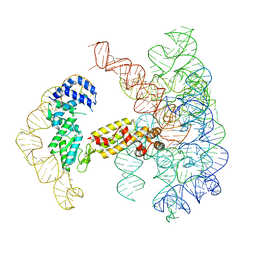 | |
7UIN
 
 | | CryoEM Structure of an Group II Intron Retroelement | | Descriptor: | AMMONIUM ION, DNA (37-MER), E.r IIC Intron, ... | | Authors: | Chung, K, Xu, L. | | Deposit date: | 2022-03-29 | | Release date: | 2022-11-23 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structures of a mobile intron retroelement poised to attack its structured DNA target
Science, 378, 2022
|
|
