3DH3
 
 | | Crystal Structure of RluF in complex with a 22 nucleotide RNA substrate | | Descriptor: | Ribosomal large subunit pseudouridine synthase F, stem loop fragment of E. Coli 23S RNA | | Authors: | Alian, A, DeGiovanni, A, Stroud, R.M, Finer-Moore, J.S. | | Deposit date: | 2008-06-16 | | Release date: | 2009-04-28 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of an RluF-RNA complex: a base-pair rearrangement is the key to selectivity of RluF for U2604 of the ribosome.
J.Mol.Biol., 388, 2009
|
|
3BT7
 
 | |
7LP1
 
 | | Structure of Nedd4L WW3 domain | | Descriptor: | E3 ubiquitin-protein ligase NEDD4-like, GLYCEROL, NITRATE ION | | Authors: | Alian, A, Alam, S.L, Thompson, T, Rheinemann, L, Sundquist, W.I. | | Deposit date: | 2021-02-11 | | Release date: | 2021-07-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Interactions between AMOT PPxY motifs and NEDD4L WW domains function in HIV-1 release.
J.Biol.Chem., 297, 2021
|
|
7LP3
 
 | | Structure of Nedd4L WW3 domain | | Descriptor: | Angiomotin, E3 ubiquitin-protein ligase NEDD4-like, SULFATE ION | | Authors: | Alian, A, Alam, S.L, Thompson, T, Rheinemann, L, Sundquist, W.I. | | Deposit date: | 2021-02-11 | | Release date: | 2021-07-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Interactions between AMOT PPxY motifs and NEDD4L WW domains function in HIV-1 release.
J.Biol.Chem., 297, 2021
|
|
7LP2
 
 | | Structure of Nedd4L WW3 domain | | Descriptor: | Angiomotin, E3 ubiquitin-protein ligase, GLYCEROL, ... | | Authors: | Alian, A, Alam, S.L, Thompson, T, Rheinemann, L, Sundquist, W.I. | | Deposit date: | 2021-02-11 | | Release date: | 2021-07-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Interactions between AMOT PPxY motifs and NEDD4L WW domains function in HIV-1 release.
J.Biol.Chem., 297, 2021
|
|
4WYV
 
 | |
5OVN
 
 | | Crystal Structure of FIV Reverse Transcriptase | | Descriptor: | POL protein | | Authors: | Galilee, M, Alian, A. | | Deposit date: | 2017-08-29 | | Release date: | 2018-01-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.942 Å) | | Cite: | The structure of FIV reverse transcriptase and its implications for non-nucleoside inhibitor resistance.
PLoS Pathog., 14, 2018
|
|
6EX9
 
 | |
2H7R
 
 | |
2H7S
 
 | | L244A mutant of Cytochrome P450cam | | Descriptor: | Cytochrome P450-cam, HEME C | | Authors: | Verras, A, Alian, A, Montellano, P.R. | | Deposit date: | 2006-06-02 | | Release date: | 2006-10-31 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Cytochrome P450 active site plasticity: attenuation of imidazole binding in cytochrome P450cam by an L244A mutation.
Protein Eng.Des.Sel., 19, 2006
|
|
2H7Q
 
 | | Cytochrome P450cam complexed with imidazole | | Descriptor: | Cytochrome P450-cam, IMIDAZOLE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Verras, A, Alian, A, Montellano, P.R. | | Deposit date: | 2006-06-02 | | Release date: | 2006-10-31 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Cytochrome P450 active site plasticity: attenuation of imidazole binding in cytochrome P450cam by an L244A mutation.
Protein Eng.Des.Sel., 19, 2006
|
|
5DCK
 
 | |
5EU7
 
 | | Crystal structure of HIV-1 integrase catalytic core in complex with Fab | | Descriptor: | FAB Heavy Chain, FAB light chain, Integrase | | Authors: | Galilee, M, Griner, S.L, Stroud, R.M, Alian, A. | | Deposit date: | 2015-11-18 | | Release date: | 2016-09-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | The Preserved HTH-Docking Cleft of HIV-1 Integrase Is Functionally Critical.
Structure, 24, 2016
|
|
4TRJ
 
 | | Crystal structure of Mycobacterium tuberculosis enoyl reductase (INHA) complexed with N-(3-bromophenyl)-1-cyclohexyl-5-oxopyrrolidine-3-carboxamide, refined with new ligand restraints | | Descriptor: | (3S)-N-(3-BROMOPHENYL)-1-CYCLOHEXYL-5-OXOPYRROLIDINE-3-CARBOXAMIDE, Enoyl-[acyl-carrier-protein] reductase [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | He, X, Alian, A, Stroud, R.M, Ortiz de Montellano, P.R. | | Deposit date: | 2014-06-17 | | Release date: | 2014-08-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Pyrrolidine carboxamides as a novel class of inhibitors of enoyl acyl carrier protein reductase from Mycobacterium tuberculosis
J. Med. Chem., 2006
|
|
4TZT
 
 | | CRYSTAL STRUCTURE OF MYCOBACTERIUM TUBERCULOSIS ENOYL REDUCTASE (INHA) COMPLEXED WITH N-(3-CHLORO-2-METHYLPHENYL)-1-CYCLOHEXYL- 5-OXOPYRROLIDINE-3-CARBOXAMIDE | | Descriptor: | (3S)-N-(3-CHLORO-2-METHYLPHENYL)-1-CYCLOHEXYL-5-OXOPYRROLIDINE-3-CARBOXAMIDE, Enoyl-[acyl-carrier-protein] reductase [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | He, X, Alian, A, Stroud, R.M, Ortiz de Montellano, P.R. | | Deposit date: | 2014-07-10 | | Release date: | 2014-08-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Pyrrolidine carboxamides as a novel class of inhibitors of enoyl acyl carrier protein reductase from Mycobacterium tuberculosis
J. Med. Chem., 2006
|
|
4U0J
 
 | | Crystal structure of Mycobacterium tuberculosis enoyl reductase (INHA) complexed with 1-CYCLOHEXYL-5-OXO-N-PHENYLPYRROLIDINE-3-CARBOXAMIDE, refined with new ligand restraints | | Descriptor: | (3S)-1-CYCLOHEXYL-5-OXO-N-PHENYLPYRROLIDINE-3-CARBOXAMIDE, Enoyl-[acyl-carrier-protein] reductase [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | He, X, Alian, A, Stroud, R.M, Ortiz de Montellano, P.R. | | Deposit date: | 2014-07-11 | | Release date: | 2014-07-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Pyrrolidine carboxamides as a novel class of inhibitors of enoyl acyl carrier protein reductase from Mycobacterium tuberculosis
J. Med. Chem., 2006
|
|
4U0K
 
 | | Crystal structure of Mycobacterium tuberculosis enoyl reductase complexed with N-(5-chloro-2-methylphenyl)-1-cyclohexyl-5-oxopyrrolidine-3-carboxamide | | Descriptor: | (3S)-N-(5-CHLORO-2-METHYLPHENYL)-1-CYCLOHEXYL-5-OXOPYRROLIDINE-3-CARBOXAMIDE, Enoyl-[acyl-carrier-protein] reductase [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | He, X, Alian, A, Stroud, R.M, Ortiz de Montellano, P.R. | | Deposit date: | 2014-07-11 | | Release date: | 2014-07-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Pyrrolidine carboxamides as a novel class of inhibitors of enoyl acyl carrier protein reductase from Mycobacterium tuberculosis
J. Med. Chem., 2006
|
|
4TZK
 
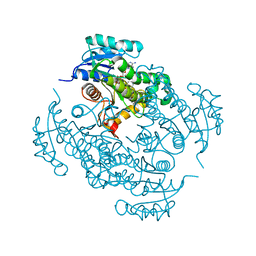 | | Crystal structure of Mycobacterium tuberculosis enoyl reductase (INHA) complexed WITH 1-CYCLOHEXYL-N-(3,5-DICHLOROPHENYL)-5-OXOPYRROLIDINE-3-CARBOXAMIDE | | Descriptor: | (3S)-1-CYCLOHEXYL-N-(3,5-DICHLOROPHENYL)-5-OXOPYRROLIDINE-3-CARBOXAMIDE, Enoyl-[acyl-carrier-protein] reductase [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | He, X, Alian, A, Stroud, R.M, Ortiz de Montellano, P.R. | | Deposit date: | 2014-07-10 | | Release date: | 2014-08-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Pyrrolidine carboxamides as a novel class of inhibitors of enoyl acyl carrier protein reductase from Mycobacterium tuberculosis
J. Med. Chem., 2006
|
|
7LP5
 
 | | Structure of Nedd4L WW3 domain | | Descriptor: | Angiomotin,E3 ubiquitin-protein ligase NEDD4-like | | Authors: | Alam, S.L, Alian, A, Thompson, T, Rheinemann, L, Volkman, B.F, Peterson, F.C, Sundquist, W.I. | | Deposit date: | 2021-02-11 | | Release date: | 2021-07-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Interactions between AMOT PPxY motifs and NEDD4L WW domains function in HIV-1 release.
J.Biol.Chem., 297, 2021
|
|
7LP4
 
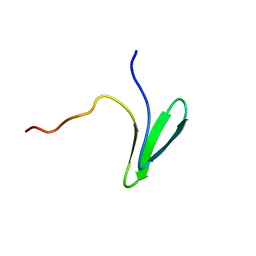 | | Structure of Nedd4L WW3 domain | | Descriptor: | E3 ubiquitin-protein ligase NEDD4-like | | Authors: | Alam, S.L, Alian, A, Thompson, T, Rheinemann, L, Volkman, B.F, Peterson, F.C, Sundquist, W.I. | | Deposit date: | 2021-02-11 | | Release date: | 2021-07-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Interactions between AMOT PPxY motifs and NEDD4L WW domains function in HIV-1 release.
J.Biol.Chem., 297, 2021
|
|
2NSD
 
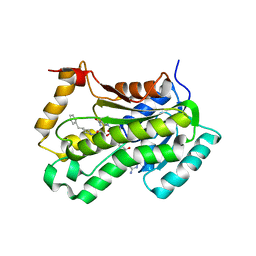 | |
4P9C
 
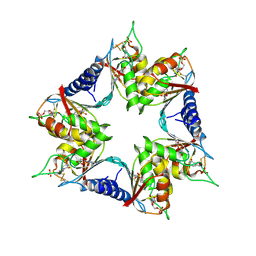 | |
4P9E
 
 | |
4P9D
 
 | |
4PA1
 
 | |
