8DTX
 
 | | Crystal structure of SARS-CoV-2 spike stem helix peptide in complex with neutralizing antibody COV89-22 | | Descriptor: | COV89-22 heavy chain, COV89-22 light chain, Spike protein S2' stem helix peptide | | Authors: | Lin, T.H, Lee, C.C.D, Wilson, I.A. | | Deposit date: | 2022-07-26 | | Release date: | 2022-11-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Rare, convergent antibodies targeting the stem helix broadly neutralize diverse betacoronaviruses.
Cell Host Microbe, 31, 2023
|
|
8DTR
 
 | | Crystal structure of SARS-CoV-2 spike stem helix peptide in complex with neutralizing antibody COV30-14 | | Descriptor: | COV30-14 heavy chain, COV30-14 light chain, Spike protein S2' stem helix peptide | | Authors: | Lee, C.C.D, Lin, T.H, Wilson, I.A. | | Deposit date: | 2022-07-26 | | Release date: | 2022-11-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Rare, convergent antibodies targeting the stem helix broadly neutralize diverse betacoronaviruses.
Cell Host Microbe, 31, 2023
|
|
7XSS
 
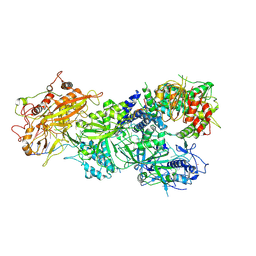 | | Structure of Craspase-CTR | | Descriptor: | CHAT domain protein, RAMP superfamily protein, RNA (34-MER), ... | | Authors: | Feng, Y, Zang, L.X. | | Deposit date: | 2022-05-15 | | Release date: | 2022-11-09 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Target RNA activates the protease activity of Craspase to confer antiviral defense.
Mol.Cell, 82, 2022
|
|
8DTT
 
 | | Crystal structure of SARS-CoV-2 spike stem helix peptide in complex with neutralizing antibody COV93-03 | | Descriptor: | COV93-03 heavy chain, COV93-03 light chain, Spike protein S2' stem helix peptide | | Authors: | Lee, C.C.D, Lin, T.H, Wilson, I.A. | | Deposit date: | 2022-07-26 | | Release date: | 2022-11-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Rare, convergent antibodies targeting the stem helix broadly neutralize diverse betacoronaviruses.
Cell Host Microbe, 31, 2023
|
|
4O2D
 
 | |
7YJ0
 
 | |
4NCE
 
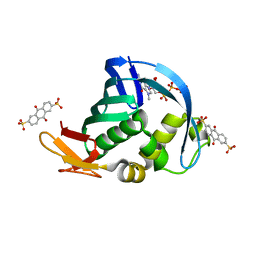 | | Influenza polymerase basic protein 2 (PB2) bound to 7-methyl-GTP | | Descriptor: | 7N-METHYL-8-HYDROGUANOSINE-5'-TRIPHOSPHATE, 9,10-dioxo-9,10-dihydroanthracene-2,6-disulfonic acid, Polymerase basic protein 2 | | Authors: | Jacobs, M.D. | | Deposit date: | 2013-10-24 | | Release date: | 2014-07-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery of a Novel, First-in-Class, Orally Bioavailable Azaindole Inhibitor (VX-787) of Influenza PB2.
J.Med.Chem., 57, 2014
|
|
6VYC
 
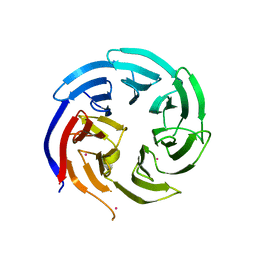 | | Crystal structure of WD-repeat domain of human WDR91 | | Descriptor: | UNKNOWN ATOM OR ION, WD repeat-containing protein 91 | | Authors: | Halabelian, L, Hutchinson, A, Li, Y, Seitova, A, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-02-26 | | Release date: | 2020-03-25 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Discovery of a First-in-Class Small-Molecule Ligand for WDR91 Using DNA-Encoded Chemical Library Selection Followed by Machine Learning.
J.Med.Chem., 66, 2023
|
|
4MUX
 
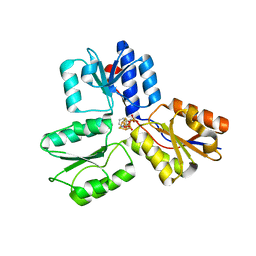 | | IspH in complex with pyridin-3-ylmethyl diphosphate | | Descriptor: | 4-hydroxy-3-methylbut-2-enyl diphosphate reductase, IRON/SULFUR CLUSTER, pyridin-3-ylmethyl trihydrogen diphosphate | | Authors: | Span, I, Eisenreich, W, Bacher, A, Oldfield, E, Groll, M. | | Deposit date: | 2013-09-23 | | Release date: | 2014-06-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Insights into the Binding of Pyridines to the Iron-Sulfur Enzyme IspH.
J.Am.Chem.Soc., 136, 2014
|
|
6W5N
 
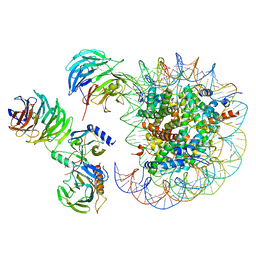 | | Cryo-EM structure of MLL1 in complex with RbBP5, WDR5, SET1, and ASH2L bound to the nucleosome (Class05) | | Descriptor: | DNA (147-MER), Histone H2A type 1, Histone H2B 1.1, ... | | Authors: | Park, S.H, Lee, Y.T, Ayoub, A, Dou, Y, Cho, U. | | Deposit date: | 2020-03-13 | | Release date: | 2021-03-31 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (6 Å) | | Cite: | Mechanism for DPY30 and ASH2L intrinsically disordered regions to modulate the MLL/SET1 activity on chromatin.
Nat Commun, 12, 2021
|
|
6W5I
 
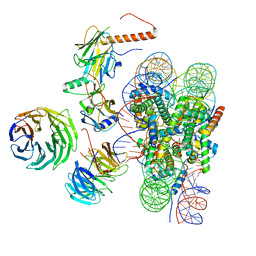 | | Cryo-EM structure of MLL1 in complex with RbBP5, WDR5, SET1, and ASH2L bound to the nucleosome (Class01) | | Descriptor: | DNA (147-MER), Histone H2A, Histone H2B 1.1, ... | | Authors: | Park, S.H, Lee, Y.T, Ayoub, A, Dou, Y, Cho, U. | | Deposit date: | 2020-03-13 | | Release date: | 2021-03-31 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (6.9 Å) | | Cite: | Mechanism for DPY30 and ASH2L intrinsically disordered regions to modulate the MLL/SET1 activity on chromatin.
Nat Commun, 12, 2021
|
|
6W5M
 
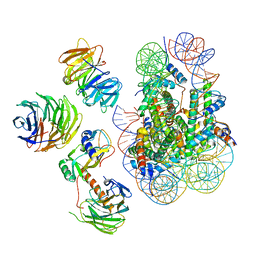 | | Cryo-EM structure of MLL1 in complex with RbBP5, WDR5, SET1, and ASH2L bound to the nucleosome (Class02) | | Descriptor: | DNA (147-MER), Histone H2A type 1, Histone H2B 1.1, ... | | Authors: | Park, S.H, Lee, Y.T, Ayoub, A, Dou, Y, Cho, U. | | Deposit date: | 2020-03-13 | | Release date: | 2021-03-31 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Mechanism for DPY30 and ASH2L intrinsically disordered regions to modulate the MLL/SET1 activity on chromatin.
Nat Commun, 12, 2021
|
|
7RPK
 
 | | Cryo-EM structure of murine Dispatched in complex with Sonic hedgehog | | Descriptor: | (2S)-3-{[(S)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-2-(hexanoyloxy)propyl hexanoate, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Asarnow, D, Wang, Q, Ding, K, Cheng, Y, Beachy, P.A. | | Deposit date: | 2021-08-03 | | Release date: | 2021-10-27 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Dispatched uses Na + flux to power release of lipid-modified Hedgehog.
Nature, 599, 2021
|
|
7RPJ
 
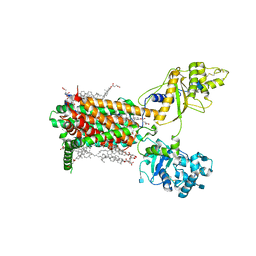 | | Cryo-EM structure of murine Dispatched NNN mutant | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL HEMISUCCINATE, Protein dispatched homolog 1 | | Authors: | Asarnow, D, Wang, Q, Ding, K, Cheng, Y, Beachy, P.A. | | Deposit date: | 2021-08-03 | | Release date: | 2021-10-27 | | Last modified: | 2021-11-24 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Dispatched uses Na + flux to power release of lipid-modified Hedgehog.
Nature, 599, 2021
|
|
7RPH
 
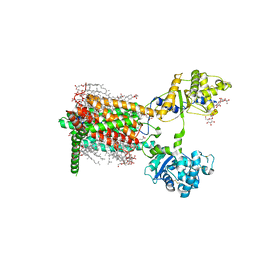 | | Cryo-EM structure of murine Dispatched 'R' conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL HEMISUCCINATE, Lauryl Maltose Neopentyl Glycol, ... | | Authors: | Asarnow, D, Wang, Q, Ding, K, Cheng, Y, Beachy, P.A. | | Deposit date: | 2021-08-03 | | Release date: | 2021-10-27 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Dispatched uses Na + flux to power release of lipid-modified Hedgehog.
Nature, 599, 2021
|
|
7RPI
 
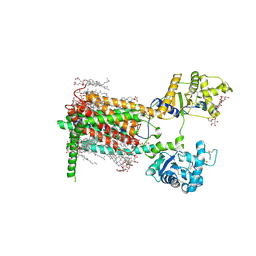 | | Cryo-EM structure of murine Dispatched 'T' conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL HEMISUCCINATE, Lauryl Maltose Neopentyl Glycol, ... | | Authors: | Asarnow, D, Wang, Q, Ding, K, Cheng, Y, Beachy, P.A. | | Deposit date: | 2021-08-03 | | Release date: | 2021-10-27 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Dispatched uses Na + flux to power release of lipid-modified Hedgehog.
Nature, 599, 2021
|
|
6JM5
 
 | | Crystal structure of TBC1D23 C terminal domain | | Descriptor: | SODIUM ION, TBC1 domain family member 23 | | Authors: | Sun, Q, Huang, W. | | Deposit date: | 2019-03-07 | | Release date: | 2019-10-16 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and functional studies of TBC1D23 C-terminal domain provide a link between endosomal trafficking and PCH.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
8ESS
 
 | | Myoglobin variant Mb-cIII complex | | Descriptor: | Myoglobin, SULFATE ION, [2,18-bis(2-carboxyethyl)-7,12-diethenyl-3,8,13,17-tetramethyl-21-(2-oxo-3-phenylpropyl)porphyrin-21-iumato(2-)-kappa~3~N~22~,N~23~,N~24~]iron(2+) | | Authors: | Bacik, J.P, Fasan, R, Ando, N. | | Deposit date: | 2022-10-14 | | Release date: | 2023-12-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Mechanistic manifold in a hemoprotein-catalyzed cyclopropanation reaction with diazoketone.
Nat Commun, 14, 2023
|
|
8ESU
 
 | | Myoglobin variant Mb-imi complex | | Descriptor: | IMIDAZOLE, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Bacik, J.P, Fasan, R, Ando, N. | | Deposit date: | 2022-10-14 | | Release date: | 2023-12-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | Mechanistic manifold in a hemoprotein-catalyzed cyclopropanation reaction with diazoketone.
Nat Commun, 14, 2023
|
|
7XYD
 
 | | Crystal structure of TMPRSS2 in complex with Nafamostat | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-carbamimidamidobenzoic acid, CALCIUM ION, ... | | Authors: | Wang, H, Liu, X, Duan, Y, Liu, X, Sun, L, Yang, H. | | Deposit date: | 2022-06-01 | | Release date: | 2023-12-06 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Structure-based discovery of dual pathway inhibitors for SARS-CoV-2 entry.
Nat Commun, 14, 2023
|
|
7Y0F
 
 | | Crystal structure of TMPRSS2 in complex with UK-371804 | | Descriptor: | 2-[(1-carbamimidamido-4-chloranyl-isoquinolin-7-yl)sulfonylamino]-2-methyl-propanoic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Wang, H, Duan, Y, Liu, X, Sun, L, Yang, H. | | Deposit date: | 2022-06-04 | | Release date: | 2023-12-06 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure-based discovery of dual pathway inhibitors for SARS-CoV-2 entry.
Nat Commun, 14, 2023
|
|
7Y0E
 
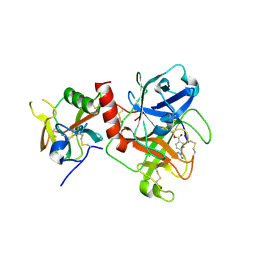 | | Crystal structure of TMPRSS2 in complex with Camostat | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-carbamimidamidobenzoic acid, CALCIUM ION, ... | | Authors: | Wang, H, Duan, Y, Liu, X, Sun, L, Yang, H. | | Deposit date: | 2022-06-04 | | Release date: | 2023-12-06 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Structure-based discovery of dual pathway inhibitors for SARS-CoV-2 entry.
Nat Commun, 14, 2023
|
|
7MLB
 
 | |
7MLI
 
 | |
7MLJ
 
 | |
