6U2D
 
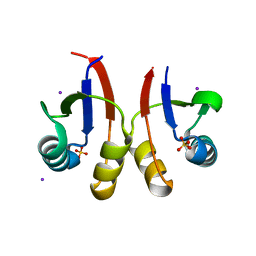 | | PmtCD peptide exporter basket domain | | Descriptor: | ABC transporter ATP-binding protein, IODIDE ION, SULFATE ION | | Authors: | Zeytuni, N, Strynadka, N.C.J. | | Deposit date: | 2019-08-19 | | Release date: | 2020-10-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Structural insight into the Staphylococcus aureus ATP-driven exporter of virulent peptide toxins
Sci Adv, 6, 2020
|
|
5XEP
 
 | | Crystal structure of BRP39, a chitinase-like protein, at 2.6 Angstorm resolution | | Descriptor: | 1,2-ETHANEDIOL, Chitinase-3-like protein 1 | | Authors: | Mohanty, A.K, Fisher, A.J, Choudhary, S, Kaushik, J.K. | | Deposit date: | 2017-04-05 | | Release date: | 2018-04-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of BRP39, a signalling glycoprotein expressed during mammary gland apoptosis.
To be published
|
|
7FJI
 
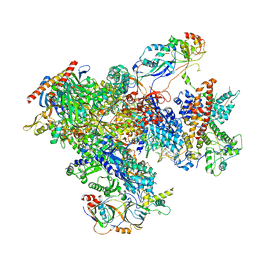 | | human Pol III elongation complex | | Descriptor: | DNA-directed RNA polymerase III subunit RPC1, DNA-directed RNA polymerase III subunit RPC10, DNA-directed RNA polymerase III subunit RPC2, ... | | Authors: | Hou, H, Xu, Y. | | Deposit date: | 2021-08-04 | | Release date: | 2021-10-27 | | Last modified: | 2021-11-10 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural insights into RNA polymerase III-mediated transcription termination through trapping poly-deoxythymidine.
Nat Commun, 12, 2021
|
|
7FJJ
 
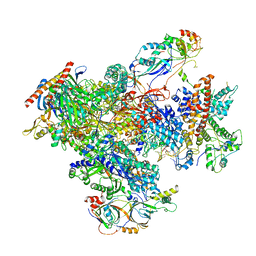 | | human Pol III pre-termination complex | | Descriptor: | DNA-directed RNA polymerase III subunit RPC1, DNA-directed RNA polymerase III subunit RPC10, DNA-directed RNA polymerase III subunit RPC2, ... | | Authors: | Hou, H, Xu, Y. | | Deposit date: | 2021-08-04 | | Release date: | 2021-10-27 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural insights into RNA polymerase III-mediated transcription termination through trapping poly-deoxythymidine.
Nat Commun, 12, 2021
|
|
5YGI
 
 | |
7JN4
 
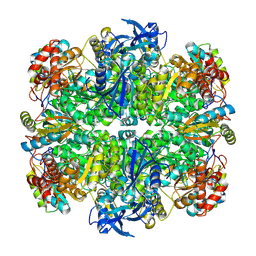 | | Rubisco in the apo state | | Descriptor: | Ribulose bisphosphate carboxylase large chain, Ribulose bisphosphate carboxylase small chain 2, chloroplastic | | Authors: | Matthies, D, Jonikas, M.C, He, S. | | Deposit date: | 2020-08-03 | | Release date: | 2020-11-18 | | Last modified: | 2020-12-23 | | Method: | ELECTRON MICROSCOPY (2.68 Å) | | Cite: | The structural basis of Rubisco phase separation in the pyrenoid.
Nat.Plants, 6, 2020
|
|
7JFO
 
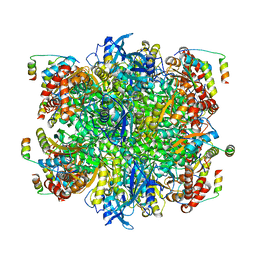 | | EPYC1(49-72)-bound Rubisco | | Descriptor: | LCI5, Ribulose bisphosphate carboxylase large chain, Ribulose bisphosphate carboxylase small chain 2, ... | | Authors: | Matthies, D, Jonikas, M.C, He, S. | | Deposit date: | 2020-07-17 | | Release date: | 2020-11-18 | | Last modified: | 2020-12-23 | | Method: | ELECTRON MICROSCOPY (2.13 Å) | | Cite: | The structural basis of Rubisco phase separation in the pyrenoid.
Nat.Plants, 6, 2020
|
|
7JSX
 
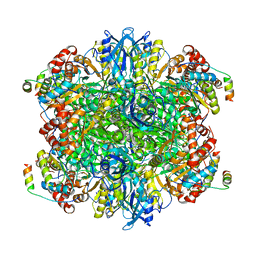 | | EPYC1(106-135) peptide-bound Rubisco | | Descriptor: | EPYC1, Ribulose bisphosphate carboxylase large chain, Ribulose bisphosphate carboxylase small chain 2, ... | | Authors: | Matthies, D, He, S, Jonikas, M.C. | | Deposit date: | 2020-08-16 | | Release date: | 2020-11-18 | | Last modified: | 2020-12-23 | | Method: | ELECTRON MICROSCOPY (2.06 Å) | | Cite: | The structural basis of Rubisco phase separation in the pyrenoid.
Nat.Plants, 6, 2020
|
|
6A9F
 
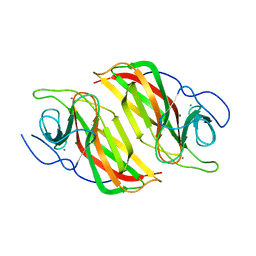 | | Crystal structure of a cyclase from Fischerella sp. TAU in complex with 4-(1H-Indol-3-yl)butan-2-one | | Descriptor: | 4-(1~{H}-indol-3-yl)butan-2-one, CALCIUM ION, GLYCEROL, ... | | Authors: | Hu, X.Y, Liu, W.D, Chen, C.C, Guo, R.T. | | Deposit date: | 2018-07-13 | | Release date: | 2018-12-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Crystal Structure of a Class of Cyclases that Catalyze the Cope Rearrangement
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
6A99
 
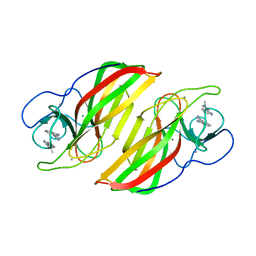 | | Crystal structure of a Stig cyclases Fisc from Fischerella sp. TAU in complex with (3Z)-3-(1-methyl-2-pyrrolidinylidene)-3H-indole | | Descriptor: | (3~{Z})-3-(1-methylpyrrolidin-2-ylidene)indole, CALCIUM ION, MAGNESIUM ION, ... | | Authors: | Hu, X.Y, Liu, W.D, Chen, C.C, Guo, R.T. | | Deposit date: | 2018-07-12 | | Release date: | 2018-12-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | The Crystal Structure of a Class of Cyclases that Catalyze the Cope Rearrangement
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
6ADU
 
 | | Crystal structure of an enzyme in complex with ligand C | | Descriptor: | (3~{Z})-3-(1-methylpyrrolidin-2-ylidene)indole, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, ... | | Authors: | Tan, X.K, Liu, W.D, Chen, C.C, Guo, R.T. | | Deposit date: | 2018-08-02 | | Release date: | 2019-08-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | The Crystal Structure of a Class of Cyclases that Catalyze the Cope Rearrangement.
Angew.Chem.Int.Ed.Engl., 57, 2018
|
|
8HDJ
 
 | |
8HER
 
 | |
8HEP
 
 | |
8HEQ
 
 | |
6B88
 
 | | E. coli LepB in complex with GNE0775 ((4S,7S,10S)-10-((S)-4-amino-2-(2-(4-(tert-butyl)phenyl)-4-methylpyrimidine-5-carboxamido)-N-methylbutanamido)-16,26-bis(2-aminoethoxy)-N-(2-iminoethyl)-7-methyl-6,9-dioxo-5,8-diaza-1,2(1,3)-dibenzenacyclodecaphane-4-carboxamide) | | Descriptor: | (8S,11S,14S)-14-{[(2S)-4-amino-2-{[2-(4-tert-butylphenyl)-4-methylpyrimidine-5-carbonyl]amino}butanoyl](methyl)amino}-3,18-bis(2-aminoethoxy)-N-[(2Z)-2-iminoethyl]-11-methyl-10,13-dioxo-9,12-diazatricyclo[13.3.1.1~2,6~]icosa-1(19),2(20),3,5,15,17-hexaene-8-carboxamide, PENTAETHYLENE GLYCOL, Signal peptidase I | | Authors: | Murray, J.M, Rouge, L. | | Deposit date: | 2017-10-05 | | Release date: | 2018-10-10 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.407 Å) | | Cite: | Optimized arylomycins are a new class of Gram-negative antibiotics.
Nature, 561, 2018
|
|
6BPD
 
 | |
6BPA
 
 | | Plasmodium vivax reticulocyte binding protein 2b (PvRBP2b) bound to monoclonal antibody 3E9 | | Descriptor: | BROMIDE ION, Monoclonal antibody 3E9 Fab heavy chain, Monoclonal antibody 3E9 Fab light chain, ... | | Authors: | Gruszczyk, J, Chan, L.J, Tham, W.H. | | Deposit date: | 2017-11-22 | | Release date: | 2018-06-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Cryo-EM structure of an essential Plasmodium vivax invasion complex.
Nature, 559, 2018
|
|
6BPE
 
 | | Plasmodium vivax reticulocyte binding protein 2b (PvRBP2b) bound to monoclonal antibody 6H1 | | Descriptor: | Monoclonal antibody 6H1 Fab heavy chain, Monoclonal antibody 6H1 Fab light chain, Reticulocyte binding protein 2, ... | | Authors: | Gruszczyk, J, Chan, L.J, Tham, W.H. | | Deposit date: | 2017-11-22 | | Release date: | 2018-06-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.34 Å) | | Cite: | Cryo-EM structure of an essential Plasmodium vivax invasion complex.
Nature, 559, 2018
|
|
6BPB
 
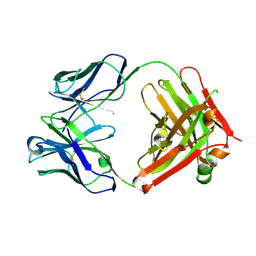 | |
6BPC
 
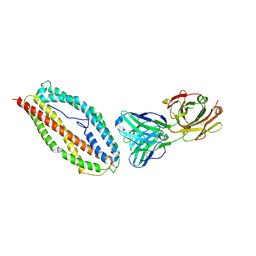 | | Plasmodium vivax reticulocyte binding protein 2b (PvRBP2b) bound to monoclonal antibody 4F7 | | Descriptor: | Monoclonal antibody 4F7 Fab heavy chain, Monoclonal antibody 4F7 Fab light chain, Reticulocyte binding protein 2, ... | | Authors: | Gruszczyk, J, Chan, L.J, Tham, W.H. | | Deposit date: | 2017-11-22 | | Release date: | 2018-06-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Cryo-EM structure of an essential Plasmodium vivax invasion complex.
Nature, 559, 2018
|
|
8XOM
 
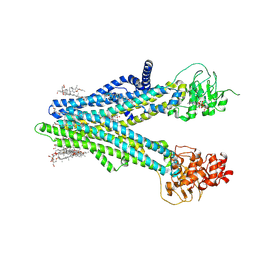 | | Cryo-EM structure of human ABCC4 in complex with ANP-bound in NBD1 and METHOTREXATE | | Descriptor: | 2-[2-[(1~{S},2~{S},4~{S},5'~{R},6~{R},7~{S},8~{R},9~{S},12~{S},13~{R},16~{S})-5',7,9,13-tetramethylspiro[5-oxapentacyclo[10.8.0.0^{2,9}.0^{4,8}.0^{13,18}]icos-18-ene-6,2'-oxane]-16-yl]oxyethyl]propane-1,3-diol, ATP-binding cassette sub-family C member 4, MAGNESIUM ION, ... | | Authors: | Zhang, P.F, Liu, Z. | | Deposit date: | 2024-01-01 | | Release date: | 2024-07-17 | | Last modified: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | The ATP-bound inward-open conformation of ABCC4 reveals asymmetric ATP binding for substrate transport.
Febs Lett., 598, 2024
|
|
8XOL
 
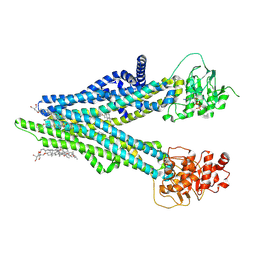 | | Cryo-EM structure of human ABCC4 with ANP bound in NBD1 | | Descriptor: | 2-[2-[(1~{S},2~{S},4~{S},5'~{R},6~{R},7~{S},8~{R},9~{S},12~{S},13~{R},16~{S})-5',7,9,13-tetramethylspiro[5-oxapentacyclo[10.8.0.0^{2,9}.0^{4,8}.0^{13,18}]icos-18-ene-6,2'-oxane]-16-yl]oxyethyl]propane-1,3-diol, ATP-binding cassette sub-family C member 4, MAGNESIUM ION, ... | | Authors: | Zhang, P.F, Liu, Z. | | Deposit date: | 2024-01-01 | | Release date: | 2024-07-17 | | Last modified: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (3.02 Å) | | Cite: | The ATP-bound inward-open conformation of ABCC4 reveals asymmetric ATP binding for substrate transport.
Febs Lett., 598, 2024
|
|
8XOK
 
 | | Cryo-EM structure of human ABCC4 | | Descriptor: | 2-[2-[(1~{S},2~{S},4~{S},5'~{R},6~{R},7~{S},8~{R},9~{S},12~{S},13~{R},16~{S})-5',7,9,13-tetramethylspiro[5-oxapentacyclo[10.8.0.0^{2,9}.0^{4,8}.0^{13,18}]icos-18-ene-6,2'-oxane]-16-yl]oxyethyl]propane-1,3-diol, ATP-binding cassette sub-family C member 4, PALMITIC ACID | | Authors: | Zhang, P.F, Liu, Z. | | Deposit date: | 2024-01-01 | | Release date: | 2024-07-24 | | Last modified: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (2.84 Å) | | Cite: | The ATP-bound inward-open conformation of ABCC4 reveals asymmetric ATP binding for substrate transport.
Febs Lett., 598, 2024
|
|
3RIK
 
 | | The acid beta-glucosidase active site exhibits plasticity in binding 3,4,5,6-tetrahydroxyazepane-based inhibitors: implications for pharmacological chaperone design for gaucher disease | | Descriptor: | (3S,4R,5R,6S)-1-(2-hydroxyethyl)azepane-3,4,5,6-tetrol, 2-acetamido-2-deoxy-beta-D-glucopyranose, Glucosylceramidase, ... | | Authors: | Orwig, S.D, Lieberman, R.L. | | Deposit date: | 2011-04-13 | | Release date: | 2012-03-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Binding of 3,4,5,6-tetrahydroxyazepanes to the acid-beta-glucosidase active site: implications for pharmacological chaperone design for Gaucher disease
Biochemistry, 50, 2011
|
|
