6DNS
 
 | | Endo-fucoidan hydrolase MfFcnA9 from glycoside hydrolase family 107 | | Descriptor: | 1,2-ETHANEDIOL, Alpha-1,4-endofucoidanase, CALCIUM ION | | Authors: | Vickers, C, Abe, K, Salama-Alber, O, Boraston, A.B. | | Deposit date: | 2018-06-07 | | Release date: | 2018-10-03 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Endo-fucoidan hydrolases from glycoside hydrolase family 107 (GH107) display structural and mechanistic similarities to alpha-l-fucosidases from GH29.
J. Biol. Chem., 293, 2018
|
|
2DIJ
 
 | | COMPLEX OF A Y195F MUTANT CGTASE FROM B. CIRCULANS STRAIN 251 COMPLEXED WITH A MALTONONAOSE INHIBITOR AT PH 9.8 OBTAINED AFTER SOAKING THE CRYSTAL WITH ACARBOSE AND MALTOHEXAOSE | | Descriptor: | 1-AMINO-2,3-DIHYDROXY-5-HYDROXYMETHYL CYCLOHEX-5-ENE, CALCIUM ION, CYCLODEXTRIN GLYCOSYLTRANSFERASE, ... | | Authors: | Strokopytov, B.V, Knegtel, R.M.A, Uitdehaag, J.C.M, Dijkstra, B.W. | | Deposit date: | 1998-05-27 | | Release date: | 1998-12-09 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of cyclodextrin glycosyltransferase complexed with a maltononaose inhibitor at 2.6 angstrom resolution. Implications for product specificity.
Biochemistry, 35, 1996
|
|
5L0U
 
 | | human POGLUT1 in complex with EGF(+) and UDP-phosphono-glucose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, EGF(+), ... | | Authors: | Li, Z, Rini, J.M. | | Deposit date: | 2016-07-28 | | Release date: | 2017-08-09 | | Last modified: | 2021-03-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis of Notch O-glucosylation and O-xylosylation by mammalian protein-O-glucosyltransferase 1 (POGLUT1).
Nat Commun, 8, 2017
|
|
5L0V
 
 | | human POGLUT1 in complex with 2F-glucose modified EGF(+) and UDP | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-deoxy-2-fluoro-beta-D-glucopyranose, ... | | Authors: | Li, Z, Rini, J.M. | | Deposit date: | 2016-07-28 | | Release date: | 2017-08-09 | | Last modified: | 2021-03-24 | | Method: | X-RAY DIFFRACTION (1.305 Å) | | Cite: | Structural basis of Notch O-glucosylation and O-xylosylation by mammalian protein-O-glucosyltransferase 1 (POGLUT1).
Nat Commun, 8, 2017
|
|
5L0S
 
 | | human POGLUT1 in complex with Factor VII EGF1 and UDP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Li, Z, Rini, J.M. | | Deposit date: | 2016-07-28 | | Release date: | 2017-08-09 | | Last modified: | 2021-03-24 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural basis of Notch O-glucosylation and O-xylosylation by mammalian protein-O-glucosyltransferase 1 (POGLUT1).
Nat Commun, 8, 2017
|
|
5L0R
 
 | | human POGLUT1 in complex with Notch1 EGF12 and UDP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Li, Z, Rini, J.M. | | Deposit date: | 2016-07-28 | | Release date: | 2017-08-09 | | Last modified: | 2021-03-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural basis of Notch O-glucosylation and O-xylosylation by mammalian protein-O-glucosyltransferase 1 (POGLUT1).
Nat Commun, 8, 2017
|
|
8GPB
 
 | |
4GPB
 
 | |
4GQR
 
 | | Human Pancreatic alpha-amylase in complex with myricetin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 3,5,7-TRIHYDROXY-2-(3,4,5-TRIHYDROXYPHENYL)-4H-CHROMEN-4-ONE, CALCIUM ION, ... | | Authors: | Williams, L.K, Brayer, G.D. | | Deposit date: | 2012-08-23 | | Release date: | 2012-10-24 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Order and disorder: differential structural impacts of myricetin and ethyl caffeate on human amylase, an antidiabetic target.
J.Med.Chem., 55, 2012
|
|
4GQQ
 
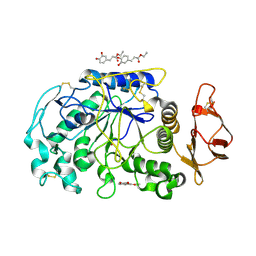 | | Human pancreatic alpha-amylase with bound ethyl caffeate | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Williams, L.K, Brayer, G.D. | | Deposit date: | 2012-08-23 | | Release date: | 2012-10-24 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Order and disorder: differential structural impacts of myricetin and ethyl caffeate on human amylase, an antidiabetic target.
J.Med.Chem., 55, 2012
|
|
4X6L
 
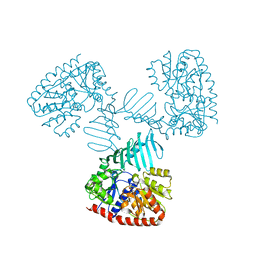 | | Crystal structure of S. aureus TarM in complex with UDP | | Descriptor: | TarM, URIDINE-5'-DIPHOSPHATE | | Authors: | Worrall, L.J, Sobhanifar, S, Gruninger, R.J, Strynadka, N.C. | | Deposit date: | 2014-12-08 | | Release date: | 2015-02-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | Structure and mechanism of Staphylococcus aureus TarM, the wall teichoic acid alpha-glycosyltransferase.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4X7P
 
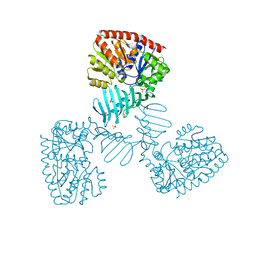 | | Crystal structure of apo S. aureus TarM | | Descriptor: | SULFATE ION, TarM | | Authors: | Worrall, L.J, Sobhanifar, S, Gruninger, R.J, Strynadka, N.C. | | Deposit date: | 2014-12-09 | | Release date: | 2015-02-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structure and mechanism of Staphylococcus aureus TarM, the wall teichoic acid alpha-glycosyltransferase.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4X7M
 
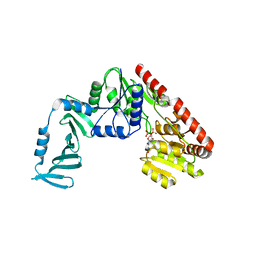 | | Crystal structure of S. aureus TarM G117R mutant in complex with UDP and UDP-GlcNAc | | Descriptor: | TarM, URIDINE-5'-DIPHOSPHATE, URIDINE-DIPHOSPHATE-N-ACETYLGLUCOSAMINE | | Authors: | Worrall, L.J, Sobhanifar, S, Strynadka, N.C. | | Deposit date: | 2014-12-09 | | Release date: | 2015-03-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure and mechanism of Staphylococcus aureus TarM, the wall teichoic acid alpha-glycosyltransferase.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4X7R
 
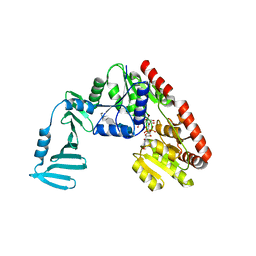 | | Crystal structure of S. aureus TarM G117R mutant in complex with Fondaparinux, alpha-GlcNAc-glycerol and UDP | | Descriptor: | (2S)-2,3-dihydroxypropyl 2-acetamido-2-deoxy-alpha-D-glucopyranoside, 2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-4)-2-deoxy-3,6-di-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-methyl 2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranoside, TarM, ... | | Authors: | Worrall, L.J, Sobhanifar, S, Strynadka, N.C. | | Deposit date: | 2014-12-09 | | Release date: | 2015-02-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure and mechanism of Staphylococcus aureus TarM, the wall teichoic acid alpha-glycosyltransferase.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4X0N
 
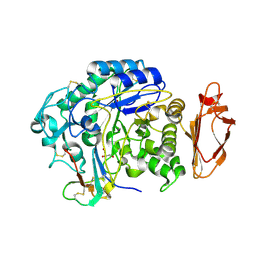 | |
6M8N
 
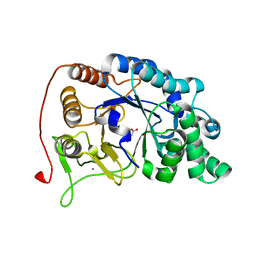 | | Endo-fucoidan hydrolase P5AFcnA from glycoside hydrolase family 107 | | Descriptor: | CALCIUM ION, MALONATE ION, P5AFcnA | | Authors: | Boraston, A.B, Vickers, C.J, Abe, K, Salama-Alber, O. | | Deposit date: | 2018-08-22 | | Release date: | 2018-10-03 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Endo-fucoidan hydrolases from glycoside hydrolase family 107 (GH107) display structural and mechanistic similarities to alpha-l-fucosidases from GH29.
J. Biol. Chem., 293, 2018
|
|
5BO8
 
 | | Structure of human sialyltransferase ST8SiaIII | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CITRIC ACID, ... | | Authors: | Volkers, G, Worrall, L, Strynadka, N.C.J. | | Deposit date: | 2015-05-27 | | Release date: | 2015-07-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of human ST8SiaIII sialyltransferase provides insight into cell-surface polysialylation.
Nat.Struct.Mol.Biol., 22, 2015
|
|
5BO6
 
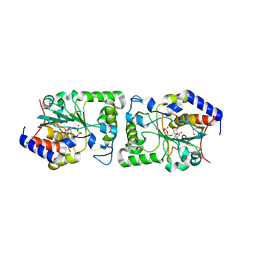 | | Structure of human sialyltransferase ST8SiaIII in complex with CDP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Volkers, G, Worrall, L, Strynadka, N.C.J. | | Deposit date: | 2015-05-27 | | Release date: | 2015-07-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Structure of human ST8SiaIII sialyltransferase provides insight into cell-surface polysialylation.
Nat.Struct.Mol.Biol., 22, 2015
|
|
5BO9
 
 | | Structure of human sialyltransferase ST8SiaIII in complex with CMP-3FNeu5Ac and Sia-6S-LacNAc | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Volkers, G, Worrall, L, Strynadka, N.C.J. | | Deposit date: | 2015-05-27 | | Release date: | 2015-07-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of human ST8SiaIII sialyltransferase provides insight into cell-surface polysialylation.
Nat.Struct.Mol.Biol., 22, 2015
|
|
5BO7
 
 | | Structure of human sialyltransferase ST8SiaIII in complex with CTP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Volkers, G, Worrall, L, Strynadka, N.C.J. | | Deposit date: | 2015-05-27 | | Release date: | 2015-07-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of human ST8SiaIII sialyltransferase provides insight into cell-surface polysialylation.
Nat.Struct.Mol.Biol., 22, 2015
|
|
5WC8
 
 | |
5WC6
 
 | |
4PBG
 
 | | 6-PHOSPHO-BETA-GALACTOSIDASE FORM-CST | | Descriptor: | 6-O-phosphono-beta-D-galactopyranose, 6-PHOSPHO-BETA-D-GALACTOSIDASE | | Authors: | Wiesmann, C, Schulz, G.E. | | Deposit date: | 1997-02-21 | | Release date: | 1997-07-23 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures and mechanism of 6-phospho-beta-galactosidase from Lactococcus lactis.
J.Mol.Biol., 269, 1997
|
|
5WD7
 
 | | Structure of a bacterial polysialyltransferase in complex with fondaparinux | | Descriptor: | 2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-4)-2-deoxy-3,6-di-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-methyl 2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranoside, SULFATE ION, SiaD | | Authors: | Worrall, L.J, Lizak, C, Strynadka, N.C.J. | | Deposit date: | 2017-07-04 | | Release date: | 2017-08-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | X-ray crystallographic structure of a bacterial polysialyltransferase provides insight into the biosynthesis of capsular polysialic acid.
Sci Rep, 7, 2017
|
|
5WCN
 
 | |
