6AS5
 
 | | CRYSTAL STRUCTURE OF PROTEIN CitE FROM MYCOBACTERIUM TUBERCULOSIS IN COMPLEX WITH MAGNESIUM, ACETOACETATE AND COENZYME A | | Descriptor: | ACETOACETIC ACID, COENZYME A, Citrate lyase subunit beta-like protein, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Wang, H, Bonanno, J.B, Carvalho, L, Almo, S.C. | | Deposit date: | 2017-08-23 | | Release date: | 2018-08-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.035 Å) | | Cite: | An essential bifunctional enzyme inMycobacterium tuberculosisfor itaconate dissimilation and leucine catabolism.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6CJ4
 
 | | CRYSTAL STRUCTURE OF PROTEIN CITE FROM MYCOBACTERIUM TUBERCULOSIS IN COMPLEX WITH MAGNESIUM AND ACETOACETATE | | Descriptor: | ACETATE ION, ACETOACETIC ACID, Citrate lyase subunit beta-like protein, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Wang, H, Bonanno, J.B, Carvalho, L, Almo, S.C. | | Deposit date: | 2018-02-26 | | Release date: | 2018-08-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | An essential bifunctional enzyme inMycobacterium tuberculosisfor itaconate dissimilation and leucine catabolism.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6CJ3
 
 | | CRYSTAL STRUCTURE OF PROTEIN CITE FROM MYCOBACTERIUM TUBERCULOSIS IN COMPLEX WITH MAGNESIUM AND PYRUVATE | | Descriptor: | ACETATE ION, Citrate lyase subunit beta-like protein, MAGNESIUM ION, ... | | Authors: | Fedorov, E.V, Fedorov, A.A, Wang, H, Bonanno, J.B, Carvalho, L, Almo, S.C. | | Deposit date: | 2018-02-26 | | Release date: | 2018-08-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | An essential bifunctional enzyme inMycobacterium tuberculosisfor itaconate dissimilation and leucine catabolism.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6CHU
 
 | | CRYSTAL STRUCTURE OF PROTEIN CITE FROM MYCOBACTERIUM TUBERCULOSIS IN COMPLEX WITH MAGNESIUM AND ACETATE | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Citrate lyase subunit beta-like protein, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Wang, H, Bonanno, J.B, Carvalho, L, Almo, S.C. | | Deposit date: | 2018-02-23 | | Release date: | 2018-08-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | An essential bifunctional enzyme inMycobacterium tuberculosisfor itaconate dissimilation and leucine catabolism.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
5TRU
 
 | | Structure of the first-in-class checkpoint inhibitor Ipilimumab bound to human CTLA-4 | | Descriptor: | Cytotoxic T-lymphocyte protein 4, Ipilimumab Fab heavy chain, Ipilimumab Fab light chain | | Authors: | Ramagopal, U.A, Liu, W, Garrett-Thomson, S.C, Yan, Q, Srinivasan, M, Wong, S.C, Bell, A, Mankikar, S, Rangan, V.S, Deshpande, S, Bonanno, J.B, Korman, A.J, Almo, S.C. | | Deposit date: | 2016-10-27 | | Release date: | 2017-05-10 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for cancer immunotherapy by the first-in-class checkpoint inhibitor ipilimumab.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
7MSJ
 
 | | The crystal structure of mouse HVEM | | Descriptor: | SULFATE ION, Tumor necrosis factor receptor superfamily member 14 | | Authors: | Liu, W, Ramagopal, U, Garrett-Thompson, S.C, Fedorov, E, Bonanno, J.B, Almo, S.C. | | Deposit date: | 2021-05-11 | | Release date: | 2021-10-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | HVEM structures and mutants reveal distinct functions of binding to LIGHT and BTLA/CD160.
J.Exp.Med., 218, 2021
|
|
7MSG
 
 | | The crystal structure of LIGHT in complex with HVEM and CD160 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CD160 antigen, soluble form,Tumor necrosis factor receptor superfamily member 14, ... | | Authors: | Liu, W, Ramagopal, U, Garrett-Thompson, S.C, Fedorov, E, Bonanno, J.B, Almo, S.C. | | Deposit date: | 2021-05-11 | | Release date: | 2021-10-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | HVEM structures and mutants reveal distinct functions of binding to LIGHT and BTLA/CD160.
J.Exp.Med., 218, 2021
|
|
5E56
 
 | | Crystal structure of mouse CTLA-4 | | Descriptor: | Cytotoxic T-lymphocyte protein 4, SODIUM ION | | Authors: | Fedorov, A.A, Fedorov, E.V, SAMANTA, D, Bonanno, J.B, Almo, S.C. | | Deposit date: | 2015-10-07 | | Release date: | 2015-10-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.504 Å) | | Cite: | Crystal structure of mouse CTLA-4
To Be Published
|
|
5DKV
 
 | | Crystal Structure of an ABC transporter Solute Binding Protein from Agrobacterium vitis(Avis_5339, TARGET EFI-511225) bound with alpha-D-Tagatopyranose | | Descriptor: | ABC transporter substrate binding protein (Ribose), alpha-D-tagatopyranose | | Authors: | Yadava, U, Al Obaidi, N.F, Morisco, L.L, Benach, J, Koss, J, Wasserman, S.R, Attonito, J.D, Scott Glenn, A, Chamala, S, Chowdhury, S, Lafleur, J, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2015-09-04 | | Release date: | 2015-09-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Crystal Structure of an ABC transporter Solute Binding Protein from Agrobacterium vitis(Avis_5339, TARGET EFI-511225) bound with alpha-D-Tagatopyranose
To be published
|
|
5E03
 
 | | Crystal structure of mouse CTLA-4 nanobody | | Descriptor: | CTLA-4 nanobody, SULFATE ION | | Authors: | Fedorov, A.A, Fedorov, E.V, Samanta, D, Bonanno, J.B, Almo, S.C. | | Deposit date: | 2015-09-28 | | Release date: | 2015-10-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.685 Å) | | Cite: | Crystal structure of mouse CTLA-4 nanobody
To Be Published
|
|
5WGG
 
 | | Structural Insights into Thioether Bond Formation in the Biosynthesis of Sactipeptides | | Descriptor: | CALCIUM ION, CteA, IRON/SULFUR CLUSTER, ... | | Authors: | Grove, T.L, Himes, P, Bowers, A, Bonanno, J.B, Almo, S.C. | | Deposit date: | 2017-07-14 | | Release date: | 2017-07-26 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.036 Å) | | Cite: | Structural Insights into Thioether Bond Formation in the Biosynthesis of Sactipeptides.
J. Am. Chem. Soc., 139, 2017
|
|
5ESR
 
 | | Crystal structure of haloalkane dehalogenase (DccA) from Caulobacter crescentus | | Descriptor: | CHLORIDE ION, COBALT (II) ION, Haloalkane dehalogenase, ... | | Authors: | Malashkevich, V.N, Toro, R, Mundorff, E.C, Almo, S.C. | | Deposit date: | 2015-11-17 | | Release date: | 2016-06-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.476 Å) | | Cite: | Biochemical characterization of two haloalkane dehalogenases: DccA from Caulobacter crescentus and DsaA from Saccharomonospora azurea.
Protein Sci., 25, 2016
|
|
5WHY
 
 | | Structural Insights into Thioether Bond Formation in the Biosynthesis of Sactipeptides | | Descriptor: | CALCIUM ION, IRON/SULFUR CLUSTER, Radical SAM domain protein, ... | | Authors: | Grove, T.L, Himes, P, Bowers, A, Bonanno, J.B, Almo, S.C. | | Deposit date: | 2017-07-18 | | Release date: | 2017-07-26 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.692 Å) | | Cite: | Structural Insights into Thioether Bond Formation in the Biosynthesis of Sactipeptides.
J. Am. Chem. Soc., 139, 2017
|
|
5DXW
 
 | | Crystal structure of mouse PD-L1 nanobody | | Descriptor: | PD-L1 nanobody, SULFATE ION | | Authors: | Fedorov, A.A, Fedorov, E.V, Samanta, D, Almo, S.C, Ploegh, H, Ingram, J, Dougan, M. | | Deposit date: | 2015-09-24 | | Release date: | 2015-10-07 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.441 Å) | | Cite: | Crystal structure of mouse PD-L1 nanobody
To Be Published
|
|
5VG6
 
 | | Crystal structure of D-isomer specific 2-hydroxyacid dehydrogenase from Xanthobacter autotrophicus Py2 in complex with NADPH and MES. | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, D-isomer specific 2-hydroxyacid dehydrogenase NAD-binding, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Lipowska, J, Shabalin, I.G, Kutner, J, Gasiorowska, O.A, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2017-04-10 | | Release date: | 2017-04-26 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of D-isomer specific 2-hydroxyacid dehydrogenase from Xanthobacter autotrophicus Py2 in complex with NADPH and MES.
to be published
|
|
5VE2
 
 | | Crystal structure of enoyl-CoA hydratase/isomerase from Pseudoalteromonas atlantica T6c at 2.3 A resolution. | | Descriptor: | DI(HYDROXYETHYL)ETHER, Enoyl-CoA hydratase, GLYCEROL, ... | | Authors: | Siuda, M.K, Shabalin, I.G, Cooper, D.R, Chapman, H.C, Tkaczuk, K.L, Bonanno, J, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2017-04-03 | | Release date: | 2017-04-19 | | Last modified: | 2019-12-04 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of enoyl-CoA hydratase/isomerase from Pseudoalteromonas atlantica T6c at 2.3 A resolution.
to be published
|
|
5HQJ
 
 | | Crystal structure of ABC transporter Solute Binding Protein B1G1H7 from Burkholderia graminis C4D1M, target EFI-511179, in complex with D-arabinose | | Descriptor: | CHLORIDE ION, Periplasmic binding protein/LacI transcriptional regulator, alpha-D-arabinopyranose | | Authors: | Roth, Y, Vetting, M.W, Al Obaidi, N.F, Toro, R, Morisco, L.L, Benach, J, Koss, J, Wasserman, S.R, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2016-01-21 | | Release date: | 2016-03-02 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure of ABC transporter Solute Binding Protein B1G1H7 from Burkholderia graminis C4D1M, target EFI-511179, in complex with D-arabinose
To be published
|
|
1K0K
 
 | |
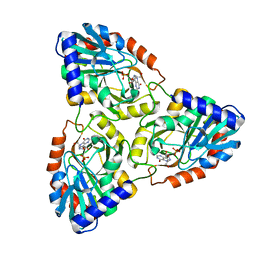
1G2P
 
 | | CRYSTAL STRUCTURE OF ADENINE PHOSPHORIBOSYLTRANSFERASE | | Descriptor: | ADENINE PHOSPHORIBOSYLTRANSFERASE 1, SULFATE ION | | Authors: | Shi, W, Tanaka, K.S.E, Almo, S.C, Schramm, V.L. | | Deposit date: | 2000-10-20 | | Release date: | 2001-12-05 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural analysis of adenine phosphoribosyltransferase from Saccharomyces cerevisiae.
Biochemistry, 40, 2001
|
|
1G2Q
 
 | |
1G2O
 
 | | CRYSTAL STRUCTURE OF PURINE NUCLEOSIDE PHOSPHORYLASE FROM MYCOBACTERIUM TUBERCULOSIS IN COMPLEX WITH A TRANSITION-STATE INHIBITOR | | Descriptor: | 1,4-DIDEOXY-4-AZA-1-(S)-(9-DEAZAHYPOXANTHIN-9-YL)-D-RIBITOL, PHOSPHATE ION, PURINE NUCLEOSIDE PHOSPHORYLASE | | Authors: | Shi, W, Basso, L.A, Tyler, P.C, Furneaux, R.H, Blanchard, J.S, Almo, S.C, Schramm, V.L. | | Deposit date: | 2000-10-20 | | Release date: | 2001-08-01 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structures of purine nucleoside phosphorylase from Mycobacterium tuberculosis in complexes with immucillin-H and its pieces.
Biochemistry, 40, 2001
|
|
1I80
 
 | | CRYSTAL STRUCTURE OF M. TUBERCULOSIS PNP IN COMPLEX WITH IMINORIBITOL, 9-DEAZAHYPOXANTHINE AND PHOSPHATE ION | | Descriptor: | 9-DEAZAHYPOXANTHINE, IMINORIBITOL, PHOSPHATE ION, ... | | Authors: | Shi, W, Basso, L.A, Tyler, P.C, Furneaux, R.H, Blanchard, J.S, Almo, S.C, Schramm, V.L. | | Deposit date: | 2001-03-12 | | Release date: | 2001-08-01 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of purine nucleoside phosphorylase from Mycobacterium tuberculosis in complexes with immucillin-H and its pieces.
Biochemistry, 40, 2001
|
|
1I85
 
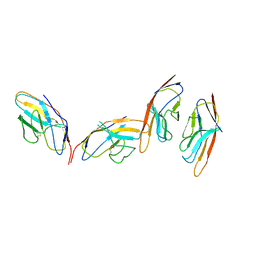 | | CRYSTAL STRUCTURE OF THE CTLA-4/B7-2 COMPLEX | | Descriptor: | CYTOTOXIC T-LYMPHOCYTE-ASSOCIATED PROTEIN 4, T LYMPHOCYTE ACTIVATION ANTIGEN CD86 | | Authors: | Schwartz, J.-C.D, Zhang, X, Fedorov, A.A, Nathenson, S.G, Almo, S.C. | | Deposit date: | 2001-03-12 | | Release date: | 2001-04-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis for co-stimulation by the human CTLA-4/B7-2 complex.
Nature, 410, 2001
|
|
1EZR
 
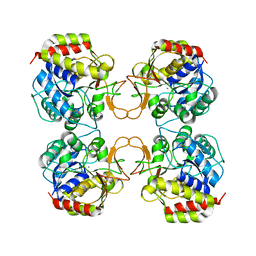 | | CRYSTAL STRUCTURE OF NUCLEOSIDE HYDROLASE FROM LEISHMANIA MAJOR | | Descriptor: | CALCIUM ION, NUCLEOSIDE HYDROLASE | | Authors: | Shi, W, Schramm, V.L, Almo, S.C. | | Deposit date: | 2000-05-11 | | Release date: | 2000-05-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Nucleoside hydrolase from Leishmania major. Cloning, expression, catalytic properties, transition state inhibitors, and the 2.5-a crystal structure.
J.Biol.Chem., 274, 1999
|
|
1INQ
 
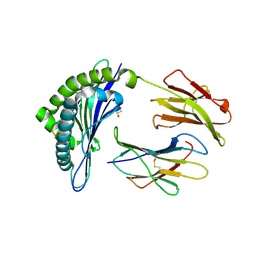 | | Structure of Minor Histocompatibility Antigen peptide, H13a, complexed to H2-Db | | Descriptor: | BETA-2 MICROGLOBULIN, DIMETHYL SULFOXIDE, H-2 CLASS I HISTOCOMPATIBILITY ANTIGEN, ... | | Authors: | Ostrov, D.A, Roden, M.M, Shi, W, Palmieri, E, Christianson, G.J, Mendoza, L, Villaflor, G, Tilley, D, Shastri, N, Grey, H, Almo, S.C, Roopenian, D, Nathenson, S.G. | | Deposit date: | 2001-05-14 | | Release date: | 2002-03-20 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | How H13 histocompatibility peptides differing by a single methyl group and lacking conventional MHC binding anchor motifs determine self-nonself discrimination.
J.Immunol., 168, 2002
|
|
