4NIE
 
 | |
5ZYQ
 
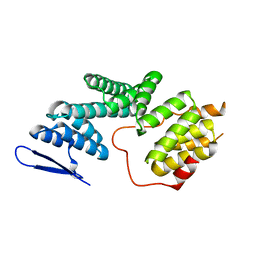 | | The Structure of Human PAF1/CTR9 complex | | Descriptor: | RNA polymerase-associated protein CTR9 homolog,RNA polymerase II-associated factor 1 homolog | | Authors: | Xie, Y, Zheng, M, Zhou, H, Long, J. | | Deposit date: | 2018-05-28 | | Release date: | 2018-09-26 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.531 Å) | | Cite: | Paf1 and Ctr9 subcomplex formation is essential for Paf1 complex assembly and functional regulation.
Nat Commun, 9, 2018
|
|
6UBS
 
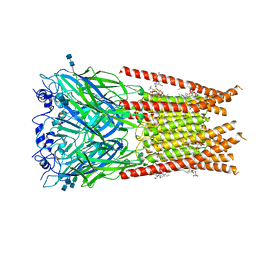 | | Full length Glycine receptor reconstituted in lipid nanodisc in Apo/Resting conformation | | Descriptor: | 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glycine receptor subunit alphaZ1, ... | | Authors: | Kumar, A, Basak, S, Chakrapani, S. | | Deposit date: | 2019-09-12 | | Release date: | 2020-07-29 | | Last modified: | 2020-08-12 | | Method: | ELECTRON MICROSCOPY (3.33 Å) | | Cite: | Mechanisms of activation and desensitization of full-length glycine receptor in lipid nanodiscs.
Nat Commun, 11, 2020
|
|
5ZYP
 
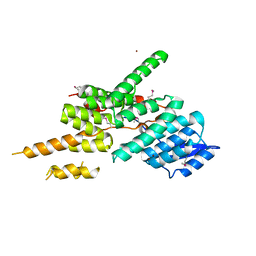 | | Structure of the Yeast Ctr9/Paf1 complex | | Descriptor: | NICKEL (II) ION, RNA polymerase-associated protein CTR9,RNA polymerase II-associated protein 1 | | Authors: | Xie, Y, Zheng, M, Zhou, H, Long, J. | | Deposit date: | 2018-05-28 | | Release date: | 2018-09-26 | | Last modified: | 2019-05-22 | | Method: | X-RAY DIFFRACTION (2.532 Å) | | Cite: | Paf1 and Ctr9 subcomplex formation is essential for Paf1 complex assembly and functional regulation.
Nat Commun, 9, 2018
|
|
7XJR
 
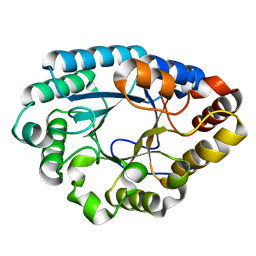 | | MLXase AlXyn26A | | Descriptor: | AlXyn26A | | Authors: | Zhang, Y.Z, Chen, X.L, Zhao, F, Yu, C.M. | | Deposit date: | 2022-04-18 | | Release date: | 2023-04-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A novel class of xylanases specifically degrade marine red algal beta 1,3/1,4-mixed-linkage xylan.
J.Biol.Chem., 299, 2023
|
|
2G2X
 
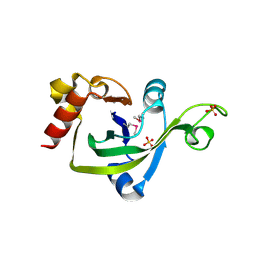 | | X-Ray Crystal Structure Protein Q88CH6 from Pseudomonas putida. Northeast Structural Genomics Consortium Target PpR72. | | Descriptor: | SULFATE ION, hypothetical protein PP5205 | | Authors: | Kuzin, A.P, Chen, Y, Abashidze, M, Acton, T, Conover, K, Janjua, H, Ma, L.-C, Ho, C.K, Cunningham, K, Montelione, G, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-02-16 | | Release date: | 2006-03-14 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural genomics reveals EVE as a new ASCH/PUA-related domain.
Proteins, 75, 2009
|
|
7YLE
 
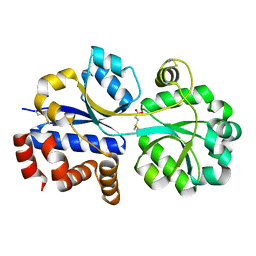 | | RnDmpX in complex with DMSP | | Descriptor: | 3-(dimethyl-lambda~4~-sulfanyl)propanoic acid, Glycine betaine/proline ABC transporter, periplasmic glycinebetaine/proline-binding protein | | Authors: | Li, C.Y. | | Deposit date: | 2022-07-26 | | Release date: | 2023-01-25 | | Last modified: | 2023-04-05 | | Method: | X-RAY DIFFRACTION (2.052 Å) | | Cite: | Ubiquitous occurrence of a dimethylsulfoniopropionate ABC transporter in abundant marine bacteria.
Isme J, 17, 2023
|
|
7CYQ
 
 | | Cryo-EM structure of an extended SARS-CoV-2 replication and transcription complex reveals an intermediate state in cap synthesis | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Helicase, MAGNESIUM ION, ... | | Authors: | Yan, L, Ge, J, Zheng, L, Zhang, Y, Gao, Y, Wang, T, Wang, H, Huang, Y, Li, M, Wang, Q, Rao, Z, Lou, Z. | | Deposit date: | 2020-09-04 | | Release date: | 2020-12-09 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | Cryo-EM Structure of an Extended SARS-CoV-2 Replication and Transcription Complex Reveals an Intermediate State in Cap Synthesis.
Cell, 184, 2021
|
|
4ZFC
 
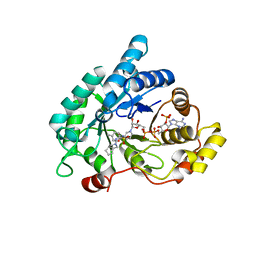 | | Crystal structure of AKR1C3 complexed with glicazide | | Descriptor: | Aldo-keto reductase family 1 member C3, N-[(3aR,6aS)-hexahydrocyclopenta[c]pyrrol-2(1H)-ylcarbamoyl]-4-methylbenzenesulfonamide, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Zhao, Y, Zhrng, X, Hu, X. | | Deposit date: | 2015-04-21 | | Release date: | 2015-11-25 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | In vitro inhibition of AKR1Cs by sulphonylureas and the structural basis
Chem.Biol.Interact., 240, 2015
|
|
7DX4
 
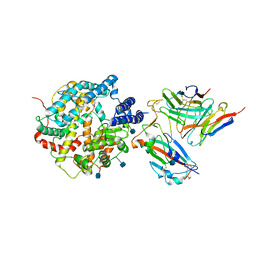 | | The structure of FC08 Fab-hA.CE2-RBD complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, Heavy chain of FC08 Fab, ... | | Authors: | Cao, L, Wang, X. | | Deposit date: | 2021-01-18 | | Release date: | 2021-04-21 | | Last modified: | 2022-02-23 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | A proof of concept for neutralizing antibody-guided vaccine design against SARS-CoV-2.
Natl Sci Rev, 8, 2021
|
|
7XS3
 
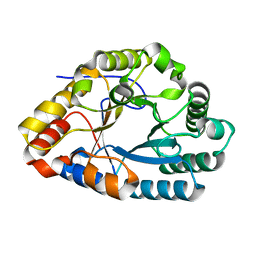 | | AlXyn26A E243A-X3X4X | | Descriptor: | AlXyn26A E243A-X3X4X, beta-D-xylopyranose-(1-3)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose | | Authors: | Zhang, Y.Z, Chen, X.L, Zhao, F, Yu, C.M. | | Deposit date: | 2022-05-12 | | Release date: | 2023-05-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A novel class of xylanases specifically degrade marine red algal beta 1,3/1,4-mixed-linkage xylan.
J.Biol.Chem., 299, 2023
|
|
6B88
 
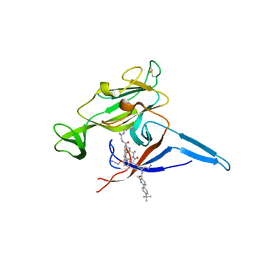 | | E. coli LepB in complex with GNE0775 ((4S,7S,10S)-10-((S)-4-amino-2-(2-(4-(tert-butyl)phenyl)-4-methylpyrimidine-5-carboxamido)-N-methylbutanamido)-16,26-bis(2-aminoethoxy)-N-(2-iminoethyl)-7-methyl-6,9-dioxo-5,8-diaza-1,2(1,3)-dibenzenacyclodecaphane-4-carboxamide) | | Descriptor: | (8S,11S,14S)-14-{[(2S)-4-amino-2-{[2-(4-tert-butylphenyl)-4-methylpyrimidine-5-carbonyl]amino}butanoyl](methyl)amino}-3,18-bis(2-aminoethoxy)-N-[(2Z)-2-iminoethyl]-11-methyl-10,13-dioxo-9,12-diazatricyclo[13.3.1.1~2,6~]icosa-1(19),2(20),3,5,15,17-hexaene-8-carboxamide, PENTAETHYLENE GLYCOL, Signal peptidase I | | Authors: | Murray, J.M, Rouge, L. | | Deposit date: | 2017-10-05 | | Release date: | 2018-10-10 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.407 Å) | | Cite: | Optimized arylomycins are a new class of Gram-negative antibiotics.
Nature, 561, 2018
|
|
6SMV
 
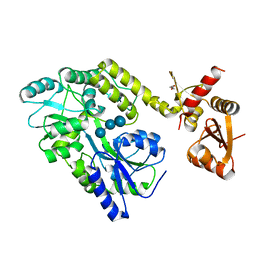 | | Structure of HPV49 E6 protein in complex with MAML1 LxxLL motif | | Descriptor: | DI(HYDROXYETHYL)ETHER, Maltose/maltodextrin-binding periplasmic protein,Protein E6,Mastermind-like protein 1, ZINC ION, ... | | Authors: | Suarez, I.P, Cousido-Siah, A, Bonhoure, A, Kostmann, C, Mitschler, A, Podjarny, A, Trave, G. | | Deposit date: | 2019-08-22 | | Release date: | 2019-09-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Cellular target recognition by HPV18 and HPV49 oncoproteins
To be published
|
|
3L95
 
 | |
7D4G
 
 | |
7EO0
 
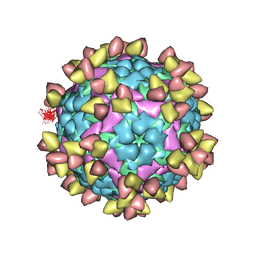 | | FOOT AND MOUTH DISEASE VIRUS O/TIBET/99-BOUND THE SINGLE CHAIN FRAGMEN ANTIBODY C4 | | Descriptor: | Ig heavy chain variable region, Ig lamda chain variable region, O/TIBET/99 VP1, ... | | Authors: | He, Y, Li, K. | | Deposit date: | 2021-04-21 | | Release date: | 2021-08-18 | | Last modified: | 2022-02-23 | | Method: | ELECTRON MICROSCOPY (3.75 Å) | | Cite: | Two Cross-Protective Antigen Sites on Foot-and-Mouth Disease Virus Serotype O Structurally Revealed by Broadly Neutralizing Antibodies from Cattle.
J.Virol., 95, 2021
|
|
6HS7
 
 | | Type VI membrane complex | | Descriptor: | ImcF-like family protein, Type VI secretion system protein VasD | | Authors: | Rapisarda, C, Fronzes, R. | | Deposit date: | 2018-09-28 | | Release date: | 2019-03-27 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | In situand high-resolution cryo-EM structure of a bacterial type VI secretion system membrane complex.
Embo J., 38, 2019
|
|
4YVX
 
 | | Crystal structure of AKR1C3 complexed with glimepiride | | Descriptor: | 3-ethyl-4-methyl-N-[2-(4-{[(cis-4-methylcyclohexyl)carbamoyl]sulfamoyl}phenyl)ethyl]-2-oxo-2,5-dihydro-1H-pyrrole-1-car boxamide, Aldo-keto reductase family 1 member C3, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Zhao, Y, Zheng, X, Zhang, H, Hu, X. | | Deposit date: | 2015-03-20 | | Release date: | 2015-11-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | In vitro inhibition of AKR1Cs by sulphonylureas and the structural basis
Chem.Biol.Interact., 240, 2015
|
|
6SIV
 
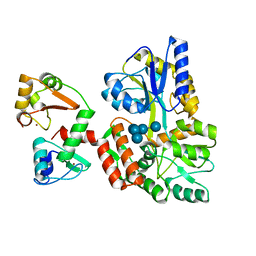 | | Structure of HPV16 E6 oncoprotein in complex with mutant IRF3 LxxLL motif | | Descriptor: | Maltose/maltodextrin-binding periplasmic protein,Interferon regulatory factor 3, Protein E6, ZINC ION, ... | | Authors: | Suarez, I.P, Cousido-Siah, A, Bonhoure, A, Mitschler, A, Podjarny, A, Trave, G. | | Deposit date: | 2019-08-12 | | Release date: | 2019-08-21 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.752 Å) | | Cite: | Deciphering the molecular and structural interaction between IRF3 and HPV16 E6
To be published
|
|
2GBS
 
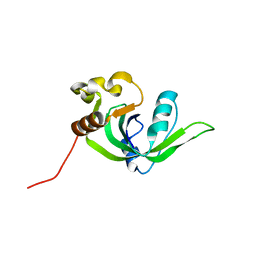 | | NMR structure of Rpa0253 from Rhodopseudomonas palustris. Northeast structural genomics consortium target RpR3 | | Descriptor: | Hypothetical protein Rpa0253 | | Authors: | Ramelot, T.A, Cort, J.R, Conover, K, Chen, Y, Ma, L.C, Ciano, M, Xiao, R, Acton, T.B, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-03-11 | | Release date: | 2006-04-11 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural genomics reveals EVE as a new ASCH/PUA-related domain.
Proteins, 75, 2009
|
|
3UA0
 
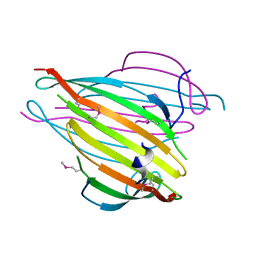 | | N-Terminal Domain of Bombyx mori Fibroin Mediates the Assembly of Silk in Response to pH Decrease | | Descriptor: | Fibroin heavy chain | | Authors: | He, Y.-X, Zhang, N.-N, Chen, B.-Y, Li, W.-F, Chen, Y.-X, Zhou, C.-Z. | | Deposit date: | 2011-10-20 | | Release date: | 2012-03-28 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | N-Terminal Domain of Bombyx mori Fibroin Mediates the Assembly of Silk in Response to pH Decrease.
J.Mol.Biol., 418, 2012
|
|
4M5E
 
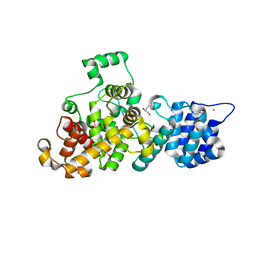 | | Tse3 structure | | Descriptor: | CADMIUM ION, CALCIUM ION, GLYCEROL, ... | | Authors: | Qian, Y. | | Deposit date: | 2013-08-08 | | Release date: | 2014-04-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Structural insights into the T6SS effector protein Tse3 and the Tse3-Tsi3 complex from Pseudomonas aeruginosa reveal a calcium-dependent membrane-binding mechanism
Mol.Microbiol., 92, 2014
|
|
6M63
 
 | | Crystal structure of a cAMP sensor G-Flamp1. | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, Chimera of Cyclic nucleotide-gated potassium channel mll3241 and Yellow fluorescent protein | | Authors: | Zhou, Z, Chen, S, Wang, L, Chu, J. | | Deposit date: | 2020-03-12 | | Release date: | 2021-09-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | A high-performance genetically encoded fluorescent indicator for in vivo cAMP imaging.
Nat Commun, 13, 2022
|
|
4G1Z
 
 | | Structural basis for the accommodation of bis- and tris-aromatic derivatives in Vitamin D Nuclear Receptor | | Descriptor: | 3-(5'-{[3,4-bis(hydroxymethyl)benzyl]oxy}-2'-ethyl-2-propylbiphenyl-4-yl)pentan-3-ol, Nuclear receptor coactivator 1, Vitamin D3 receptor A | | Authors: | Ciesielski, F, Sato, Y, Moras, D, Rochel, N. | | Deposit date: | 2012-07-11 | | Release date: | 2012-09-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for the accommodation of bis- and tris-aromatic derivatives in vitamin d nuclear receptor.
J.Med.Chem., 55, 2012
|
|
4G2H
 
 | | Structural basis for the accommodation of bis- and tris-aromatic derivatives in Vitamin D Nuclear Receptor | | Descriptor: | (3E,5E)-6-(3-{2-[3,4-bis(hydroxymethyl)phenyl]ethyl}phenyl)-1,1,1-trifluoro-2-(trifluoromethyl)octa-3,5-dien-2-ol, Nuclear receptor coactivator 1, Vitamin D3 receptor A | | Authors: | Ciesielski, F, Sato, Y, Moras, D, Rochel, N. | | Deposit date: | 2012-07-12 | | Release date: | 2012-09-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for the accommodation of bis- and tris-aromatic derivatives in vitamin d nuclear receptor.
J.Med.Chem., 55, 2012
|
|
