2WO9
 
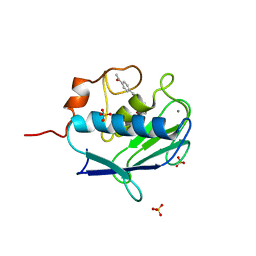 | | MMP12 complex with a beta hydroxy carboxylic acid | | Descriptor: | (3S)-5-(4'-ACETYLBIPHENYL-4-YL)-3-HYDROXYPENTANOIC ACID, CALCIUM ION, GLYCEROL, ... | | Authors: | Holmes, I.P, Gaines, S, Watson, S.P, Lorthioir, O, Walker, A, Baddeley, S.J, Herbert, S, Egan, D, Convery, M.A, Singh, O.M.P, Gross, J.W, Strelow, J.M, Smith, R.H, Amour, A.J, Brown, D, Martin, S.L. | | Deposit date: | 2009-07-22 | | Release date: | 2009-09-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Identification of Beta-Hydroxy Carboxylic Acids as Selective Mmp-12 Inhibitors.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
2WOA
 
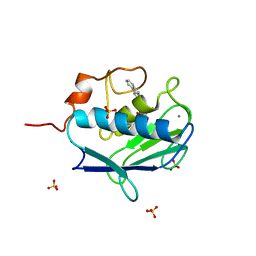 | | MMP12 complex with a beta hydroxy carboxylic acid | | Descriptor: | (3S)-5-(9H-FLUOREN-2-YL)-3-HYDROXYPENTANOIC ACID, CALCIUM ION, GLYCEROL, ... | | Authors: | Holmes, I.P, Gaines, S, Watson, S.P, Lorthioir, O, Walker, A, Baddeley, S.J, Herbert, S, Egan, D, Convery, M.A, Singh, O.M.P, Gross, J.W, Strelow, J.M, Smith, R.H, Amour, A.J, Brown, D, Martin, S.L. | | Deposit date: | 2009-07-22 | | Release date: | 2009-09-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Identification of Beta-Hydroxy Carboxylic Acids as Selective Mmp-12 Inhibitors.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
2WO8
 
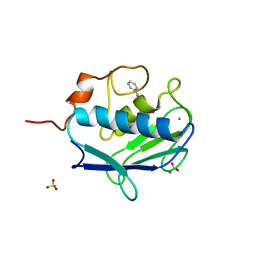 | | MMP12 complex with a beta hydroxy carboxylic acid | | Descriptor: | (3S)-5-biphenyl-4-yl-3-hydroxypentanoic acid, CALCIUM ION, GLYCEROL, ... | | Authors: | Holmes, I.P, Gaines, S, Watson, S.P, Lorthioir, O, Walker, A, Baddeley, S.J, Herbert, S, Egan, D, Convery, M.A, Singh, O.M.P, Gross, J.W, Strelow, J.M, Smith, R.H, Amour, A.J, Brown, D, Martin, S.L. | | Deposit date: | 2009-07-22 | | Release date: | 2009-09-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Identification of Beta-Hydroxy Carboxylic Acids as Selective Mmp-12 Inhibitors.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
6RIO
 
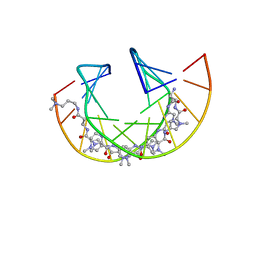 | | Imidazole Polyamide-DNA complex NMR structure (5'-CGATGTACATCG-3') | | Descriptor: | 3-[3-[[4-[[4-[[4-[[4-[[(2~{R})-2-azaniumyl-4-[[1-methyl-4-[[1-methyl-4-[[1-methyl-4-[(1-methylimidazol-2-yl)carbonylamino]pyrrol-2-yl]carbonylamino]pyrrol-2-yl]carbonylamino]pyrrol-2-yl]carbonylamino]butanoyl]amino]-1-methyl-imidazol-2-yl]carbonylamino]-1-methyl-pyrrol-2-yl]carbonylamino]-1-methyl-pyrrol-2-yl]carbonylamino]-1-methyl-pyrrol-2-yl]carbonylamino]propanoylamino]propyl-dimethyl-azanium, DNA (5'-(*(DC5)P*GP*AP*TP*GP*TP*AP*CP*AP*TP*CP*(DG3))-3') | | Authors: | Padroni, G, Withers, J.M, Taladriz-Sender, A, Reichenbach, L.F, Parkinson, J.A, Burley, G.A. | | Deposit date: | 2019-04-24 | | Release date: | 2019-06-12 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Sequence-Selective Minor Groove Recognition of a DNA Duplex Containing Synthetic Genetic Components.
J.Am.Chem.Soc., 141, 2019
|
|
2X8F
 
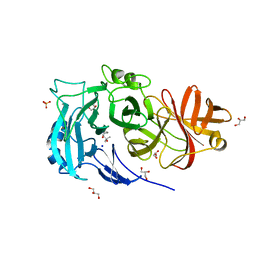 | | Native structure of Endo-1,5-alpha-L-arabinanases from Bacillus subtilis | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | de Sanctis, D, Inacio, J.M, Lindley, P.F, de Sa-Nogueira, I, Bento, I. | | Deposit date: | 2010-03-09 | | Release date: | 2011-03-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | New Evidence for the Role of Calcium in the Glycosidase Reaction of Gh43 Arabinanases.
FEBS J., 277, 2010
|
|
2Z8P
 
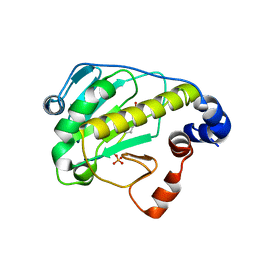 | | Structural basis for the catalytic mechanism of phosphothreonine lyase | | Descriptor: | (GLY)(GLU)(ALA)(TPO)(VAL)(PTR)(ALA), 27.5 kDa virulence protein | | Authors: | Chen, L, Wang, H, Gu, L, Huang, N, Zhou, J.M, Chai, J. | | Deposit date: | 2007-09-07 | | Release date: | 2007-12-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for the catalytic mechanism of phosphothreonine lyase.
Nat.Struct.Mol.Biol., 15, 2008
|
|
6RVM
 
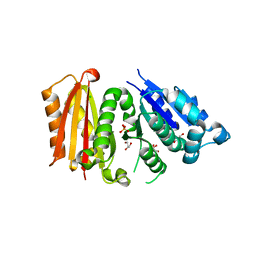 | | Cell division protein FtsZ from Staphylococcus aureus, apo form | | Descriptor: | CHLORIDE ION, Cell division protein FtsZ, GLYCEROL, ... | | Authors: | Fernandez-Tornero, C, Andreu, J.M, Canosa-Valls, A.J. | | Deposit date: | 2019-05-31 | | Release date: | 2020-02-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.155 Å) | | Cite: | Nucleotide-induced folding of cell division protein FtsZ from Staphylococcus aureus.
Febs J., 287, 2020
|
|
2ZIX
 
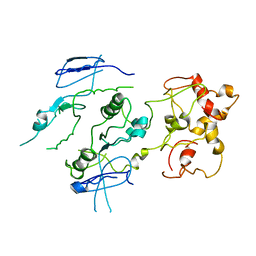 | | Crystal structure of the Mus81-Eme1 complex | | Descriptor: | Crossover junction endonuclease EME1, Crossover junction endonuclease MUS81 | | Authors: | Chang, J.H, Kim, J.J, Choi, J.M, Lee, J.H, Cho, Y. | | Deposit date: | 2008-02-25 | | Release date: | 2008-04-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystal structure of the Mus81-Eme1 complex
Genes Dev., 22, 2008
|
|
2XD9
 
 | | STRUCTURE OF HELICOBACTER PYLORI TYPE II DEHYDROQUINASE IN COMPLEX WITH INHIBITOR COMPOUND (4R,6R,7S)-4,6,7-Trihydroxy-2-((E)-prop-1- enyl)-4,5,6,7-tetrahydrobenzo(b)thiophene-4-carboxylic acid | | Descriptor: | (4R,6R,7S)-4,6,7-TRIHYDROXY-2-[(1E)-PROP-1-EN-1-YL]-4,5,6,7-TETRAHYDRO-1-BENZOTHIOPHENE-4-CARBOXYLIC ACID, 3-DEHYDROQUINATE DEHYDRATASE | | Authors: | Paz, S, Tizon, L, Otero, J.M, Llamas-Saiz, A.L, Fox, G.C, van Raaij, M.J, Lamb, H, Hawkins, A.R, Lapthorn, A.J, Castedo, L, Gonzalez-Bello, C. | | Deposit date: | 2010-04-30 | | Release date: | 2010-11-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Tetrahydrobenzothiophene derivatives: conformationally restricted inhibitors of type II dehydroquinase.
ChemMedChem, 6, 2011
|
|
2XFA
 
 | | Crystal structure of Plasmodium berghei actin depolymerization factor 2 | | Descriptor: | ACTIN DEPOLYMERIZATION FACTOR 2 | | Authors: | Singh, B.K, Sattler, J.M, Huttu, J, Chatterjee, M, Schueler, H, Kursula, I. | | Deposit date: | 2010-05-21 | | Release date: | 2011-06-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structures Explain Functional Differences in the Two Actin Depolymerization Factors of the Malaria Parasite.
J.Biol.Chem., 286, 2011
|
|
2XB9
 
 | | Structure of Helicobacter pylori type II dehydroquinase in complex with inhibitor compound (2R)-2-(4-methoxybenzyl)-3-dehydroquinic acid | | Descriptor: | (1R,2R,4S,5R)-1,4,5-TRIHYDROXY-2-(4-METHOXYBENZYL)-3-OXOCYCLOHEXANECARBOXYLIC ACID, 3-DEHYDROQUINATE DEHYDRATASE, CITRIC ACID | | Authors: | Otero, J.M, Tizon, L, Llamas-Saiz, A.L, Fox, G.C, Gonzalez-Bello, C, van Raaij, M.J. | | Deposit date: | 2010-04-08 | | Release date: | 2010-09-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Understanding the Key Factors that Control the Inhibition of Type II Dehydroquinase by (2R)-2- Benzyl-3-Dehydroquinic Acids.
Chemmedchem, 5, 2010
|
|
6SKF
 
 | | Cryo-EM Structure of T. kodakarensis 70S ribosome | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Matzov, D, Sas-Chen, A, Thomas, J.M, Santangelo, T, Meier, J.L, Schwartz, S, Shalev-Benami, M. | | Deposit date: | 2019-08-15 | | Release date: | 2020-07-29 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | Dynamic RNA acetylation revealed by quantitative cross-evolutionary mapping.
Nature, 583, 2020
|
|
2WD1
 
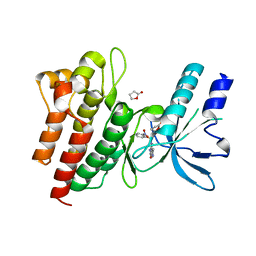 | | Human c-Met Kinase in complex with azaindole inhibitor | | Descriptor: | 1-[(2-NITROPHENYL)SULFONYL]-1H-PYRROLO[3,2-B]PYRIDINE-6-CARBOXAMIDE, GAMMA-BUTYROLACTONE, HEPATOCYTE GROWTH FACTOR RECEPTOR | | Authors: | Porter, J, Lumb, S, Franklin, R.J, Gascon-Simorte, J.M, Calmiano, M, Le Riche, K, Lallemand, B, Keyaerts, J, Edwards, H, Maloney, A, Delgado, J, King, L, Foley, A, Lecomte, F, Reuberson, J, Meier, C, Batchelor, M. | | Deposit date: | 2009-03-19 | | Release date: | 2009-04-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of 4-Azaindoles as Novel Inhibitors of C- met Kinase.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
2WLZ
 
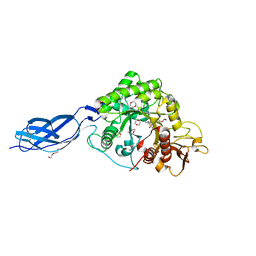 | | Chitinase A from Serratia marcescens ATCC990 in complex with Chitobio- thiazoline. | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 3AR,5R,6S,7R,7AR-5-HYDROXYMETHYL-2-METHYL-5,6,7,7A-TETRAHYDRO-3AH-PYRANO[3,2-D]THIAZOLE-6,7-DIOL, ... | | Authors: | Taylor, E.J, Dennis, R.J, Macdonald, J.M, Tarling, C.A, Knapp, S, Withers, S.G, Davies, G.J. | | Deposit date: | 2009-06-29 | | Release date: | 2010-03-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Chitinase Inhibition by Chitobiose and Chitotriose Thiazolines.
Angew.Chem.Int.Ed.Engl., 49, 2010
|
|
6R2F
 
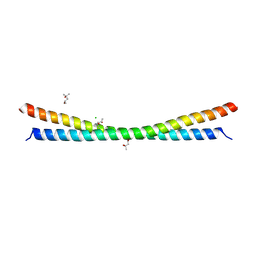 | |
2X6A
 
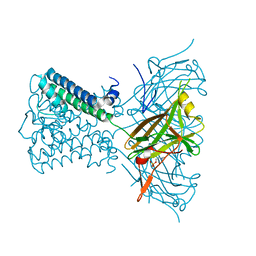 | | Potassium Channel from Magnetospirillum Magnetotacticum | | Descriptor: | ATP-SENSITIVE INWARD RECTIFIER POTASSIUM CHANNEL 10, PHOSPHOCHOLINE, POTASSIUM ION | | Authors: | Clarke, O.B, Caputo, A.T, Hill, A.P, Vandenberg, J.I, Smith, B.J, Gulbis, J.M. | | Deposit date: | 2010-02-15 | | Release date: | 2010-06-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Domain Reorientation and Rotation of an Intracellular Assembly Regulate Conduction in Kir Potassium Channels.
Cell(Cambridge,Mass.), 141, 2010
|
|
2ZCG
 
 | |
2ZL6
 
 | | Atomic resolution structural characterization of recognition of histo-blood group antigens by Norwalk virus | | Descriptor: | 58 kd capsid protein, ACETATE ION, MAGNESIUM ION, ... | | Authors: | Choi, J.M, Huston, A.M, Estes, M.K, Prasad, B.V.V. | | Deposit date: | 2008-04-02 | | Release date: | 2008-07-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Atomic resolution structural characterization of recognition of histo-blood group antigens by Norwalk virus
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
6PW8
 
 | |
6PXN
 
 | |
2ZIW
 
 | | Crystal structure of the Mus81-Eme1 complex | | Descriptor: | Crossover junction endonuclease EME1, Mus81 protein | | Authors: | Chang, J.H, Kim, J.J, Choi, J.M, Lee, J.H, Cho, Y. | | Deposit date: | 2008-02-25 | | Release date: | 2008-04-29 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the Mus81-Eme1 complex
Genes Dev., 22, 2008
|
|
2ZJP
 
 | | Thiopeptide antibiotic Nosiheptide bound to the large ribosomal subunit of Deinococcus radiodurans | | Descriptor: | 4-(hydroxymethyl)-3-methyl-1H-indole-2-carboxylic acid, 50S RIBOSOMAL PROTEIN L11, 50S RIBOSOMAL PROTEIN L13, ... | | Authors: | Harms, J.M, Wilson, D.N, Schluenzen, F, Connell, S.R, Stachelhaus, T, Zaborowska, Z, Spahn, C.M.T, Fucini, P. | | Deposit date: | 2008-03-07 | | Release date: | 2008-06-17 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Translational Regulation Via L11: Molecular Switches on the Ribosome Turned on and Off by Thiostrepton and Micrococcin.
Mol.Cell, 30, 2008
|
|
6SI9
 
 | | FtsZ-refold | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Cell division protein FtsZ, ... | | Authors: | Fernandez-Tornero, C, Andreu, J.M, Ruiz, F.M. | | Deposit date: | 2019-08-09 | | Release date: | 2020-02-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Nucleotide-induced folding of cell division protein FtsZ from Staphylococcus aureus.
Febs J., 287, 2020
|
|
6SOR
 
 | |
2ZJR
 
 | | Refined native structure of the large ribosomal subunit (50S) from Deinococcus radiodurans | | Descriptor: | 50S ribosomal protein L11, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | Authors: | Harms, J.M, Wilson, D.N, Schluenzen, F, Connell, S.R, Stachelhaus, T, Zaborowska, Z, Spahn, C.M.T, Fucini, P. | | Deposit date: | 2008-03-08 | | Release date: | 2008-06-17 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Translational regulation via L11: molecular switches on the ribosome turned on and off by thiostrepton and micrococcin.
Mol.Cell, 30, 2008
|
|
