7U1Y
 
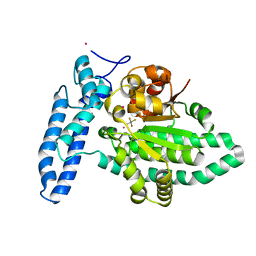 | | Structure of SPAC806.04c protein from fission yeast bound to AlF4 and Co2+ | | Descriptor: | COBALT (II) ION, Damage-control phosphatase SPAC806.04c, POTASSIUM ION, ... | | Authors: | Jacewicz, A, Sanchez, A.M, Shuman, S. | | Deposit date: | 2022-02-22 | | Release date: | 2022-06-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Fission yeast Duf89 and Duf8901 are cobalt/nickel-dependent phosphatase-pyrophosphatases that act via a covalent aspartyl-phosphate intermediate.
J.Biol.Chem., 298, 2022
|
|
6KLW
 
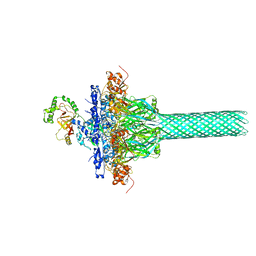 | | Complex structure of Iota toxin enzymatic component (Ia) and binding component (Ib) pore with long stem | | Descriptor: | CALCIUM ION, Iota toxin component Ia, Iota toxin component Ib | | Authors: | Yoshida, T, Yamada, T, Kawamoto, A, Mitsuoka, K, Iwasaki, K, Tsuge, H. | | Deposit date: | 2019-07-30 | | Release date: | 2020-01-15 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Cryo-EM structures reveal translocational unfolding in the clostridial binary iota toxin complex.
Nat.Struct.Mol.Biol., 27, 2020
|
|
5TVI
 
 | |
3NQ1
 
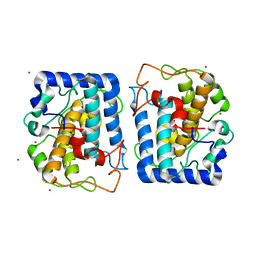 | | Crystal Structure of Tyrosinase from Bacillus megaterium in complex with inhibitor kojic acid | | Descriptor: | 5-HYDROXY-2-(HYDROXYMETHYL)-4H-PYRAN-4-ONE, COPPER (II) ION, Tyrosinase, ... | | Authors: | Sendovski, M, Kanteev, M, Adir, N, Fishman, A. | | Deposit date: | 2010-06-29 | | Release date: | 2010-11-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | First structures of an active bacterial tyrosinase reveal copper plasticity.
J.Mol.Biol., 405, 2011
|
|
3NQ5
 
 | | Crystal Structure of Tyrosinase from Bacillus megaterium R209H mutant | | Descriptor: | COPPER (II) ION, Tyrosinase, ZINC ION | | Authors: | Sendovski, M, Kanteev, M, Adir, N, Fishman, A. | | Deposit date: | 2010-06-29 | | Release date: | 2010-11-17 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | First structures of an active bacterial tyrosinase reveal copper plasticity.
J.Mol.Biol., 405, 2011
|
|
7U1V
 
 | | Structure of SPAC806.04c protein from fission yeast covalently bound to BeF3 | | Descriptor: | Damage-control phosphatase SPAC806.04c, NICKEL (II) ION, PHOSPHATE ION, ... | | Authors: | Jacewicz, A, Sanchez, A.M, Shuman, S. | | Deposit date: | 2022-02-22 | | Release date: | 2022-06-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Fission yeast Duf89 and Duf8901 are cobalt/nickel-dependent phosphatase-pyrophosphatases that act via a covalent aspartyl-phosphate intermediate.
J.Biol.Chem., 298, 2022
|
|
3JCA
 
 | | Core model of the Mouse Mammary Tumor Virus intasome | | Descriptor: | 5'-D(*AP*AP*TP*GP*CP*CP*GP*CP*AP*GP*TP*CP*GP*GP*CP*CP*GP*AP*CP*CP*TP*G)-3', 5'-D(*CP*AP*GP*GP*TP*CP*GP*GP*CP*CP*GP*AP*CP*TP*GP*CP*GP*GP*CP*A)-3', Integrase, ... | | Authors: | Lyumkis, D.L, Ballandras-Colas, A, Brown, M, Cook, N.J, Dewdney, T.G, Demeler, B, Cherepanov, P, Engelman, A.N. | | Deposit date: | 2015-11-24 | | Release date: | 2016-02-17 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Cryo-EM reveals a novel octameric integrase structure for betaretroviral intasome function.
Nature, 530, 2016
|
|
2IVP
 
 | | Structure of UP1 protein | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, FE (II) ION, O-SIALOGLYCOPROTEIN ENDOPEPTIDASE | | Authors: | Hecker, A, Leulliot, N, Graille, M, Dorlet, P, Quevillon-Cheruel, S, Ulryck, N, Van Tilbeurgh, H, Forterre, P. | | Deposit date: | 2006-06-14 | | Release date: | 2007-07-31 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | An Archaeal Orthologue of the Universal Protein Kae1 is an Iron Metalloprotein which Exhibits Atypical DNA-Binding Properties and Apurinic-Endonuclease Activity in Vitro.
Nucleic Acids Res., 35, 2007
|
|
7FCR
 
 | |
7FCS
 
 | |
7U1X
 
 | |
6C27
 
 | | SAM-III riboswitch ON-state | | Descriptor: | COBALT HEXAMMINE(III), SAM-III riboswitch | | Authors: | Grigg, J.C, Price, I.R, Ke, A. | | Deposit date: | 2018-01-07 | | Release date: | 2019-01-09 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3.601 Å) | | Cite: | Evidence for two-tiered conformation selection in the SAM-III riboswitch
To Be Published
|
|
5V54
 
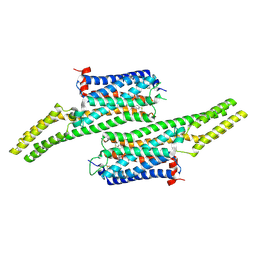 | | Crystal structure of 5-HT1B receptor in complex with methiothepin | | Descriptor: | 1-methyl-4-[(5~{S})-3-methylsulfanyl-5,6-dihydrobenzo[b][1]benzothiepin-5-yl]piperazine, 5-hydroxytryptamine receptor 1B,OB-1 fused 5-HT1b receptor,5-hydroxytryptamine receptor 1B | | Authors: | Yin, W.C, Zhou, X.E, Yang, D, de Waal, P, Wang, M.T, Dai, A, Cai, X, Huang, C.Y, Liu, P, Yin, Y, Liu, B, Caffrey, M, Melcher, K, Xu, Y, Wang, M.W, Xu, H.E, Jiang, Y. | | Deposit date: | 2017-03-13 | | Release date: | 2018-02-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | A common antagonistic mechanism for class A GPCRs revealed by the structure of the human 5-HT1B serotonin receptor bound to an antagonist
Cell Discov, 2018
|
|
1YFL
 
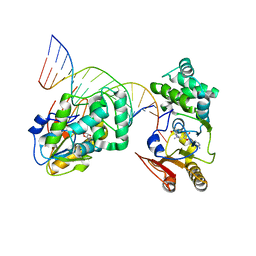 | | T4Dam in Complex with Sinefungin and 16-mer Oligonucleotide Showing Semi-specific and Specific Contact and Flipped Base | | Descriptor: | 5'-D(P*TP*CP*AP*CP*AP*GP*GP*AP*TP*CP*CP*TP*GP*TP*GP*A)-3', DNA adenine methylase, SINEFUNGIN | | Authors: | Horton, J.R, Liebert, K, Hattman, S, Jeltsch, A, Cheng, X. | | Deposit date: | 2005-01-03 | | Release date: | 2005-05-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | Transition from Nonspecific to Specific DNA Interactions along the Substrate-Recognition Pathway of Dam Methyltransferase.
Cell(Cambridge,Mass.), 121, 2005
|
|
5V0I
 
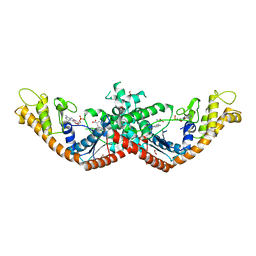 | | Crystal Structure of Tryptophanyl-tRNA Synthetase from Escherichia coli Complexed with AMP and Tryptophan | | Descriptor: | ADENOSINE MONOPHOSPHATE, FORMIC ACID, TRYPTOPHAN, ... | | Authors: | Maltseva, N, Kim, Y, Mulligan, R, Grimshaw, S.G, Joachimiak, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-02-28 | | Release date: | 2017-03-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of Tryptophanyl-tRNA Synthetase from Escherichia coli Complexed with AMP and Tryptophan
To Be Published
|
|
6C76
 
 | |
5V7P
 
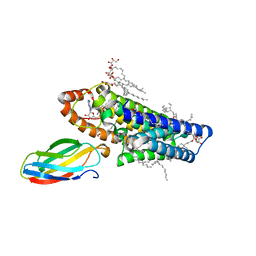 | | Atomic structure of the eukaryotic intramembrane Ras methyltransferase ICMT (isoprenylcysteine carboxyl methyltransferase), in complex with a monobody | | Descriptor: | DECANE, Protein-S-isoprenylcysteine O-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Long, S.B, Diver, M.M, Pedi, L, Koide, A, Koide, S. | | Deposit date: | 2017-03-20 | | Release date: | 2018-01-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Atomic structure of the eukaryotic intramembrane RAS methyltransferase ICMT.
Nature, 553, 2018
|
|
7TXH
 
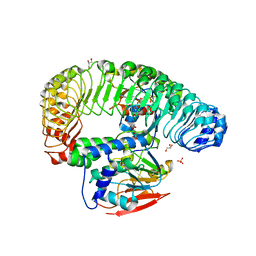 | | Human MRas Q71R in complex with human Shoc2 LRR domain M173I and human PP1Ca | | Descriptor: | GLYCEROL, Leucine-rich repeat protein SHOC-2, MAGNESIUM ION, ... | | Authors: | Hauseman, Z.J, Viscomi, J, Dhembi, A, Clark, K, King, D.A, Fodor, M. | | Deposit date: | 2022-02-09 | | Release date: | 2022-06-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of the MRAS-SHOC2-PP1C phosphatase complex.
Nature, 609, 2022
|
|
5V8F
 
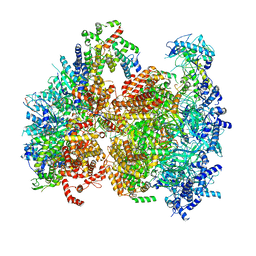 | | Structural basis of MCM2-7 replicative helicase loading by ORC-Cdc6 and Cdt1 | | Descriptor: | Cell division control protein 6, Cell division cycle protein CDT1, DNA (39-MER), ... | | Authors: | Yuan, Z, Riera, A, Bai, L, Sun, J, Spanos, C, Chen, Z.A, Barbon, M, Rappsilber, J, Stillman, B, Speck, C, Li, H. | | Deposit date: | 2017-03-21 | | Release date: | 2017-05-10 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural basis of Mcm2-7 replicative helicase loading by ORC-Cdc6 and Cdt1.
Nat. Struct. Mol. Biol., 24, 2017
|
|
5RE9
 
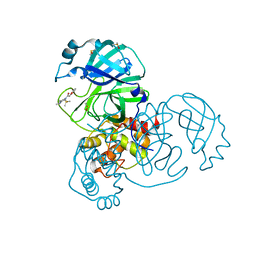 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 main protease in complex with Z2856434836 | | Descriptor: | 2-(4-methylphenoxy)-1-(4-methylpiperazin-4-ium-1-yl)ethanone, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Owen, C.D, Douangamath, A, Lukacik, P, Powell, A.J, Strain-Damerell, C.M, Resnick, E, Krojer, T, Gehrtz, P, Wild, C, Aimon, A, Brandao-Neto, J, Carbery, A, Dunnett, L, Skyner, R, Snee, M, London, N, Walsh, M.A, von Delft, F. | | Deposit date: | 2020-03-15 | | Release date: | 2020-03-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Crystallographic and electrophilic fragment screening of the SARS-CoV-2 main protease.
Nat Commun, 11, 2020
|
|
7TYG
 
 | |
5REP
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 main protease in complex with PCM-0102201 | | Descriptor: | 1-{4-[(2,6-difluorophenyl)sulfonyl]piperazin-1-yl}ethan-1-one, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Owen, C.D, Douangamath, A, Lukacik, P, Powell, A.J, Strain-Damerell, C.M, Resnick, E, Krojer, T, Gehrtz, P, Wild, C, Aimon, A, Brandao-Neto, J, Carbery, A, Dunnett, L, Skyner, R, Snee, M, London, N, Walsh, M.A, von Delft, F. | | Deposit date: | 2020-03-15 | | Release date: | 2020-03-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Crystallographic and electrophilic fragment screening of the SARS-CoV-2 main protease.
Nat Commun, 11, 2020
|
|
5R80
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 main protease in complex with Z18197050 | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, methyl 4-sulfamoylbenzoate | | Authors: | Fearon, D, Powell, A.J, Douangamath, A, Owen, C.D, Wild, C, Krojer, T, Lukacik, P, Strain-Damerell, C.M, Walsh, M.A, von Delft, F. | | Deposit date: | 2020-03-03 | | Release date: | 2020-03-11 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystallographic and electrophilic fragment screening of the SARS-CoV-2 main protease.
Nat Commun, 11, 2020
|
|
6CA0
 
 | | Cryo-EM structure of E. coli RNAP sigma70 open complex | | Descriptor: | DNA (35-MER), DNA (45-MER), DNA (5'-D(P*GP*CP*CP*GP*CP*GP*TP*CP*AP*GP*A)-3'), ... | | Authors: | Narayanan, A, Vago, F, Li, K, Qayyum, M.Z, Yernool, D, Jiang, W, Murakami, K.S. | | Deposit date: | 2018-01-29 | | Release date: | 2018-02-28 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (5.75 Å) | | Cite: | Cryo-EM structure ofEscherichia colisigma70RNA polymerase and promoter DNA complex revealed a role of sigma non-conserved region during the open complex formation.
J. Biol. Chem., 293, 2018
|
|
6JZS
 
 | | Structure of the Manganese Protoporphyrin IX-Reconstituted CYP102A1 Haem Domain with N-Abietoyl-L-Tryptophan in complex with Pyridine | | Descriptor: | (2S)-2-[[(1R,4aR,4bR,10aR)-1,4a-dimethyl-7-propan-2-yl-2,3,4,4b,5,6,10,10a-octahydrophenanthren-1-yl]carbonylamino]-3-( 1H-indol-3-yl)propanoic acid, Bifunctional cytochrome P450/NADPH--P450 reductase, GLYCEROL, ... | | Authors: | Stanfield, J.K, Omura, K, Matsumoto, A, Kasai, C, Sugimoto, H, Shiro, Y, Watanabe, Y, Shoji, O. | | Deposit date: | 2019-05-03 | | Release date: | 2020-03-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Crystals in Minutes: Instant On-Site Microcrystallisation of Various Flavours of the CYP102A1 (P450BM3) Haem Domain.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
