2MBC
 
 | | Solution Structure of human holo-PRL-3 in complex with vanadate | | Descriptor: | Protein tyrosine phosphatase type IVA 3 | | Authors: | Jeong, K, Kang, D, Kim, J, Shin, S, Jin, B, Lee, C, Kim, E, Jeon, Y.H, Kim, Y. | | Deposit date: | 2013-07-29 | | Release date: | 2013-10-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure and backbone dynamics of vanadate-bound PRL-3: comparison of 15N nuclear magnetic resonance relaxation profiles of free and vanadate-bound PRL-3.
Biochemistry, 53, 2014
|
|
5KOZ
 
 | | Structure function studies of R. palustris RubisCO (K192C mutant; CABP-bound) | | Descriptor: | 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE, CARBONATE ION, MAGNESIUM ION, ... | | Authors: | Arbing, M.A, North, J.A, Satagopan, S, Tabita, F.R. | | Deposit date: | 2016-07-01 | | Release date: | 2017-07-05 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure function studies of R. palustris RubisCO.
To Be Published
|
|
3OGI
 
 | | Crystal structure of the Mycobacterium tuberculosis H37Rv EsxOP complex (Rv2346c-Rv2347c) | | Descriptor: | Putative ESAT-6-like protein 6, Putative ESAT-6-like protein 7 | | Authors: | Arbing, M.A, Chan, S, Zhou, T.T, Ahn, C, Harris, L, Kuo, E, Sawaya, M.R, Cascio, D, Eisenberg, D, Integrated Center for Structure and Function Innovation (ISFI), TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2010-08-16 | | Release date: | 2010-08-25 | | Last modified: | 2014-05-21 | | Method: | X-RAY DIFFRACTION (2.549 Å) | | Cite: | Heterologous expression of mycobacterial Esx complexes in Escherichia coli for structural studies is facilitated by the use of maltose binding protein fusions.
Plos One, 8, 2013
|
|
3UID
 
 | | Crystal Structure of Protein Ms6760 from Mycobacterium smegmatis | | Descriptor: | Putative uncharacterized protein | | Authors: | Bajaj, R.A, Miallau, L, Cascio, D, Arbing, M, Eisenberg, D, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2011-11-04 | | Release date: | 2011-11-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.571 Å) | | Cite: | Crystal structure of the toxin Msmeg_6760, the structural homolog of Mycobacterium tuberculosis Rv2035, a novel type II toxin involved in the hypoxic response.
Acta Crystallogr F Struct Biol Commun, 72, 2016
|
|
4GZR
 
 | | Crystal structure of the Mycobacterium tuberculosis H37Rv EsxOP (Rv2346c-Rv2347c) complex in space group C2221 | | Descriptor: | ESAT-6-like protein 6, ESAT-6-like protein 7, SULFATE ION | | Authors: | Arbing, M.A, Chan, S, He, Q, Harris, L, Zhou, T.T, Kuo, E, Ahn, C.J, Eisenberg, D, TB Structural Genomics Consortium (TBSGC), Integrated Center for Structure and Function Innovation (ISFI) | | Deposit date: | 2012-09-06 | | Release date: | 2012-09-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.553 Å) | | Cite: | Heterologous expression of mycobacterial Esx complexes in Escherichia coli for structural studies is facilitated by the use of maltose binding protein fusions.
Plos One, 8, 2013
|
|
4I0X
 
 | | Crystal structure of the Mycobacterum abscessus EsxEF (Mab_3112-Mab_3113) complex | | Descriptor: | BETA-MERCAPTOETHANOL, ESAT-6-like protein MAB_3112, ESAT-6-like protein MAB_3113, ... | | Authors: | Arbing, M.A, Chan, S, Lu, J, Kuo, E, Harris, L, Eisenberg, D, Integrated Center for Structure and Function Innovation (ISFI), TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2012-11-19 | | Release date: | 2013-01-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.956 Å) | | Cite: | Heterologous expression of mycobacterial Esx complexes in Escherichia coli for structural studies is facilitated by the use of maltose binding protein fusions.
Plos One, 8, 2013
|
|
5HK4
 
 | | Structure function studies of R. palustris RubisCO (A47V-M331A mutant) | | Descriptor: | 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE, MAGNESIUM ION, Ribulose bisphosphate carboxylase | | Authors: | Arbing, M.A, Shin, A, Cascio, D, Satagopan, S, North, J.A, Tabita, F.R. | | Deposit date: | 2016-01-13 | | Release date: | 2017-01-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure function studies of R. palustris RubisCO.
To Be Published
|
|
5HQM
 
 | | Structure function studies of R. palustris RubisCO (R. palustris/R. rubrum chimera) | | Descriptor: | 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE, MAGNESIUM ION, POTASSIUM ION, ... | | Authors: | Arbing, M.A, Shin, A, Cascio, D, Satagopan, S, Tabita, F.R. | | Deposit date: | 2016-01-21 | | Release date: | 2017-01-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure function studies of R. palustris RubisCO.
To Be Published
|
|
2NCX
 
 | |
2NCY
 
 | | Solution structure of pseudin-2 analog (Ps-P) | | Descriptor: | Pseudin-2 | | Authors: | Jeon, D, Kim, J, Shin, A, Kim, Y. | | Deposit date: | 2016-04-18 | | Release date: | 2017-04-26 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Optimum Balance between the Cationicity and Structural Component for Bacterial Cell Selectivity and Anti-inflammatory activities of Pseudin-2 and its Analogs
To be Published
|
|
7NWK
 
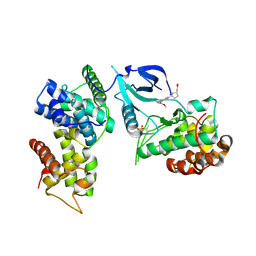 | | Crystal structure of CDK9-Cyclin T1 bound by compound 6 | | Descriptor: | Cyclin-T1, Cyclin-dependent kinase 9, N-((1R,3R)-3-(7-(4-fluoro-2-methoxyphenyl)-3H-imidazo[4,5-b]pyridin-2-yl)cyclopentyl)acetamide | | Authors: | Collie, G.W, Ferguson, A.D. | | Deposit date: | 2021-03-16 | | Release date: | 2021-10-27 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Discovery of a Series of 7-Azaindoles as Potent and Highly Selective CDK9 Inhibitors for Transient Target Engagement.
J.Med.Chem., 64, 2021
|
|
5JRF
 
 | | Crystal structure of the light-driven sodium pump KR2 bound with iodide ions | | Descriptor: | EICOSANE, IODIDE ION, Sodium pumping rhodopsin | | Authors: | Melnikov, I, Polovinkin, V, Kovalev, K, Shevchenko, V, Gushchin, I, Popov, A, Gordeliy, V. | | Deposit date: | 2016-05-06 | | Release date: | 2017-05-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Fast iodide-SAD phasing for high-throughput membrane protein structure determination.
Sci Adv, 3, 2017
|
|
5JSI
 
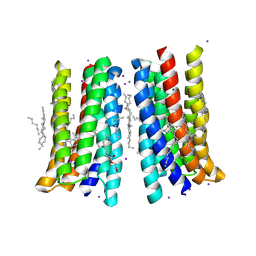 | | Structure of membrane protein | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Bacteriorhodopsin, EICOSANE, ... | | Authors: | Melnikov, I, Polovinkin, V, Kovalev, K, Shevchenko, V, Gushchin, I, Popov, A, Gordeliy, V. | | Deposit date: | 2016-05-08 | | Release date: | 2017-05-31 | | Last modified: | 2023-03-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Fast iodide-SAD phasing for high-throughput membrane protein structure determination.
Sci Adv, 3, 2017
|
|
8IMD
 
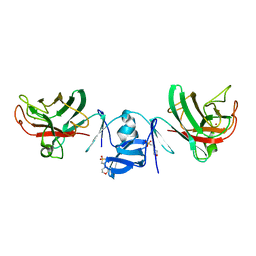 | | Crystal structure of Cu/Zn Superoxide dismutase from Paenibacillus lautus | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, COPPER (II) ION, Cu/Zn-Superoxide dismutase, ... | | Authors: | Narikiyo, S, Furukawa, Y, Akutsu, M. | | Deposit date: | 2023-03-06 | | Release date: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Characterization of a novel cysteine-less Cu/Zn-superoxide dismutase in Paenibacillus lautus missing a conserved disulfide bond.
J.Biol.Chem., 299, 2023
|
|
7AVB
 
 | |
7AVA
 
 | |
6WI6
 
 | | Crystal structure of plantacyclin B21AG | | Descriptor: | MALONATE ION, Plantacyclin B21AG | | Authors: | Smith, A.T, Gor, M.C, Vezina, B, McMahon, R, King, G, Panjikar, S, Rehm, B, Martin, J. | | Deposit date: | 2020-04-08 | | Release date: | 2021-01-06 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure and site-directed mutagenesis of circular bacteriocin plantacyclin B21AG reveals cationic and aromatic residues important for antimicrobial activity.
Sci Rep, 10, 2020
|
|
1GAI
 
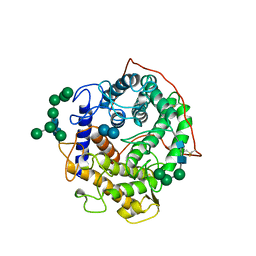 | | GLUCOAMYLASE-471 COMPLEXED WITH D-GLUCO-DIHYDROACARBOSE | | Descriptor: | 4,6-dideoxy-4-{[(1S,2S,3S,4R,5R)-2,3,4-trihydroxy-5-(hydroxymethyl)cyclohexyl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, GLUCOAMYLASE-471, alpha-D-mannopyranose, ... | | Authors: | Aleshin, A.E, Stoffer, B, Firsov, L.M, Svensson, B, Honzatko, R.B. | | Deposit date: | 1996-03-06 | | Release date: | 1996-08-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystallographic complexes of glucoamylase with maltooligosaccharide analogs: relationship of stereochemical distortions at the nonreducing end to the catalytic mechanism.
Biochemistry, 35, 1996
|
|
1GAH
 
 | | GLUCOAMYLASE-471 COMPLEXED WITH ACARBOSE | | Descriptor: | 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, GLUCOAMYLASE-471, alpha-D-mannopyranose, ... | | Authors: | Aleshin, A.E, Stoffer, B, Firsov, L.M, Svensson, B, Honzatko, R.B. | | Deposit date: | 1996-03-06 | | Release date: | 1996-08-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystallographic complexes of glucoamylase with maltooligosaccharide analogs: relationship of stereochemical distortions at the nonreducing end to the catalytic mechanism.
Biochemistry, 35, 1996
|
|
8AMO
 
 | | Crystal structure of M. tuberculosis CYP143 | | Descriptor: | CHLORIDE ION, GLYCEROL, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Bukhdruker, S, Varaksa, T, Grudo, A, Marin, E, Kapranov, I, Shevtsov, M, Gilep, A, Strushkevich, N, Borshchevskiy, V. | | Deposit date: | 2022-08-03 | | Release date: | 2023-02-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural insights into 3Fe-4S ferredoxins diversity in M. tuberculosis highlighted by a first redox complex with P450.
Front Mol Biosci, 9, 2022
|
|
8AMQ
 
 | | Crystal structure of the complex CYP143-FdxE from M. tuberculosis | | Descriptor: | FE3-S4 CLUSTER, NICKEL (II) ION, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Bukhdruker, S, Varaksa, T, Smolskaya, S, Marin, E, Kapranov, I, Kovalev, K, Gilep, A, Strushkevich, N, Borshchevskiy, V. | | Deposit date: | 2022-08-03 | | Release date: | 2023-02-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural insights into 3Fe-4S ferredoxins diversity in M. tuberculosis highlighted by a first redox complex with P450.
Front Mol Biosci, 9, 2022
|
|
7ZB9
 
 | | Crystal structure of CYP124 in complex with inhibitor carbethoxyhexyl imidazole in the absence of glycerol (NoCryo) | | Descriptor: | CHLORIDE ION, CYP124 in complex with inhibitor carbethoxyhexyl imidazole, MAGNESIUM ION, ... | | Authors: | Bukhdruker, S, Varaksa, T, Marin, E, Gilep, A, Strushkevich, N, Borshchevskiy, V. | | Deposit date: | 2022-03-23 | | Release date: | 2023-01-11 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Structural insights into the effects of glycerol on ligand binding to cytochrome P450.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8PKY
 
 | |
8PM1
 
 | |
6T0L
 
 | | Crystal structure of CYP124 in complex with inhibitor compound 5' | | Descriptor: | CHLORIDE ION, CYP124 in complex with inhibitor compound 5', DI(HYDROXYETHYL)ETHER, ... | | Authors: | Bukhdruker, S, Marin, E, Varaksa, T, Gilep, A, Strushkevich, N, Borshchevskiy, V. | | Deposit date: | 2019-10-03 | | Release date: | 2020-10-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Metabolic Fate of Human Immunoactive Sterols in Mycobacterium tuberculosis.
J.Mol.Biol., 433, 2021
|
|
