8C6P
 
 | | Fragment screening hit I bound to endothiapepsin | | Descriptor: | 4-[(2-azanyl-4-methyl-1,3-thiazol-5-yl)methyl]benzenecarbonitrile, Endothiapepsin, GLYCEROL | | Authors: | Wiese, J.N, Buehrmann, M, Mueller, M.P, Rauh, D. | | Deposit date: | 2023-01-12 | | Release date: | 2023-05-24 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Fragtory: Pharmacophore-Focused Design, Synthesis, and Evaluation of an sp 3 -Enriched Fragment Library.
J.Med.Chem., 66, 2023
|
|
8C6Q
 
 | | Fragment screening hit II bound to endothiapepsin | | Descriptor: | 3-(4-methylimidazol-1-yl)-5-(trifluoromethyl)aniline, Endothiapepsin, GLYCEROL | | Authors: | Wiese, J.N, Buehrmann, M, Mueller, M.P, Rauh, D. | | Deposit date: | 2023-01-12 | | Release date: | 2023-05-24 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Fragtory: Pharmacophore-Focused Design, Synthesis, and Evaluation of an sp 3 -Enriched Fragment Library.
J.Med.Chem., 66, 2023
|
|
8C70
 
 | | Pyrrolidine fragment 1 bound to endothiapepsin | | Descriptor: | (3~{R},4~{R})-4-[4-[(5-bromanylpyridin-3-yl)oxymethyl]-1,2,3-triazol-1-yl]pyrrolidin-3-ol, Endothiapepsin | | Authors: | Wiese, J.N, Buehrmann, M, Mueller, M.P, Rauh, D. | | Deposit date: | 2023-01-12 | | Release date: | 2023-05-24 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Fragtory: Pharmacophore-Focused Design, Synthesis, and Evaluation of an sp 3 -Enriched Fragment Library.
J.Med.Chem., 66, 2023
|
|
8C71
 
 | | Pyrrolidine fragment 5b bound to endothiapepsin | | Descriptor: | (3~{R},4~{R})-4-(3,4-dihydro-1~{H}-isoquinolin-2-yl)pyrrolidin-3-ol, Endothiapepsin, GLYCEROL | | Authors: | Wiese, J.N, Buehrmann, M, Mueller, M.P, Rauh, D. | | Deposit date: | 2023-01-12 | | Release date: | 2023-05-24 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Fragtory: Pharmacophore-Focused Design, Synthesis, and Evaluation of an sp 3 -Enriched Fragment Library.
J.Med.Chem., 66, 2023
|
|
8C72
 
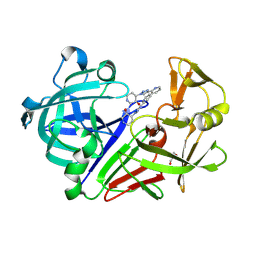 | | Pyrrolidine fragment 10b bound to endothiapepsin | | Descriptor: | (3~{S},4~{S})-4-(4-pyridin-2-yl-1,2,3-triazol-1-yl)piperidin-3-ol, Endothiapepsin, GLYCEROL | | Authors: | Wiese, J.N, Buehrmann, M, Mueller, M.P, Rauh, D. | | Deposit date: | 2023-01-12 | | Release date: | 2023-05-24 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Fragtory: Pharmacophore-Focused Design, Synthesis, and Evaluation of an sp 3 -Enriched Fragment Library.
J.Med.Chem., 66, 2023
|
|
8C74
 
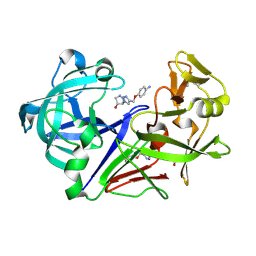 | | Pyrrolidine fragment 10d bound to endothiapepsin | | Descriptor: | (3~{R},4~{R})-4-[4-[(4-azanylphenoxy)methyl]-1,2,3-triazol-1-yl]pyrrolidin-3-ol, Endothiapepsin, GLYCEROL | | Authors: | Wiese, J.N, Buehrmann, M, Mueller, M.P, Rauh, D. | | Deposit date: | 2023-01-12 | | Release date: | 2023-05-24 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Fragtory: Pharmacophore-Focused Design, Synthesis, and Evaluation of an sp 3 -Enriched Fragment Library.
J.Med.Chem., 66, 2023
|
|
8C0Z
 
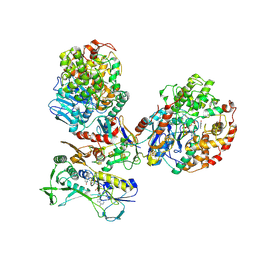 | | CryoEM structure of a tungsten-containing aldehyde oxidoreductase from Aromatoleum aromaticum | | Descriptor: | Aldehyde:ferredoxin oxidoreductase,tungsten-containing, BENZOIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Winiarska, A, Ramirez-Amador, F, Hege, D, Gemmecker, Y, Prinz, S, Hochberg, G, Heider, J, Szaleniec, M, Schuller, J.M. | | Deposit date: | 2022-12-19 | | Release date: | 2023-05-31 | | Last modified: | 2023-06-14 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | A bacterial tungsten-containing aldehyde oxidoreductase forms an enzymatic decorated protein nanowire.
Sci Adv, 9, 2023
|
|
8C6S
 
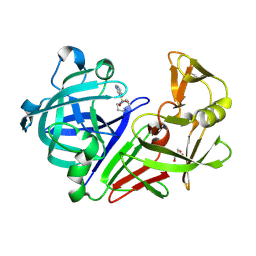 | | Fragment screening hit III bound to endothiapepsin | | Descriptor: | 4-(1,4-diazepan-1-ylsulfonyl)isoquinoline, Endothiapepsin, GLYCEROL | | Authors: | Wiese, J.N, Buehrmann, M, Mueller, M.P, Rauh, D. | | Deposit date: | 2023-01-12 | | Release date: | 2023-05-24 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Fragtory: Pharmacophore-Focused Design, Synthesis, and Evaluation of an sp 3 -Enriched Fragment Library.
J.Med.Chem., 66, 2023
|
|
8C6T
 
 | | Fragment screening hit IV bound to endothiapepsin | | Descriptor: | 1-[3,5-bis(chloranyl)phenoxy]propan-2-amine, Endothiapepsin, GLYCEROL | | Authors: | Wiese, J.N, Buehrmann, M, Mueller, M.P, Rauh, D. | | Deposit date: | 2023-01-12 | | Release date: | 2023-05-24 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Fragtory: Pharmacophore-Focused Design, Synthesis, and Evaluation of an sp 3 -Enriched Fragment Library.
J.Med.Chem., 66, 2023
|
|
6R8M
 
 | | Complex of rice blast (Magnaporthe oryzae) effector protein AVR-PikE with an engineered HMA domain of Pikp-1 from rice (Oryza sativa) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, AVR-Pik protein, CHLORIDE ION, ... | | Authors: | De la Concepcion, J.C, Franceschetti, M, Banfield, M.J. | | Deposit date: | 2019-04-02 | | Release date: | 2019-04-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Protein engineering expands the effector recognition profile of a rice NLR immune receptor.
Elife, 8, 2019
|
|
6R8K
 
 | |
8CR1
 
 | | Homo sapiens Get1/Get2 heterotetramer in complex with a Get3 dimer | | Descriptor: | ATPase ASNA1, Guided entry of tail-anchored proteins factor CAMLG,Guided entry of tail-anchored proteins factor 1,GET2-GET1, ZINC ION | | Authors: | McDowell, M.A, Heimes, M, Wild, K, Sinning, I. | | Deposit date: | 2023-03-07 | | Release date: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | The GET insertase exhibits conformational plasticity and induces membrane thinning.
Nat Commun, 14, 2023
|
|
6S6Z
 
 | | Structure of beta-Galactosidase from Thermotoga maritima | | Descriptor: | Beta-galactosidase, MAGNESIUM ION | | Authors: | Miguez-Amil, S, Jimenez-Ortega, E, Ramirez Escudero, M, Sanz-Aparicio, J, Fernandez-Leiro, R. | | Deposit date: | 2019-07-04 | | Release date: | 2020-03-18 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2 Å) | | Cite: | The cryo-EM Structure ofThermotoga maritimabeta-Galactosidase: Quaternary Structure Guides Protein Engineering.
Acs Chem.Biol., 15, 2020
|
|
8C3K
 
 | |
7SK4
 
 | | Cryo-EM structure of ACKR3 in complex with chemokine N-terminal mutant CXCL12_LRHQ, an intracellular Fab, and an extracellular Fab | | Descriptor: | Atypical chemokine receptor 3, CHOLESTEROL, CID24 Fab heavy chain, ... | | Authors: | Yen, Y.C, Schafer, C.T, Gustavsson, M, Handel, T.M, Tesmer, J.J.G. | | Deposit date: | 2021-10-19 | | Release date: | 2022-07-27 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structures of atypical chemokine receptor 3 reveal the basis for its promiscuity and signaling bias.
Sci Adv, 8, 2022
|
|
7SK6
 
 | | Cryo-EM structure of human ACKR3 in complex with chemokine N-terminal mutant CXCL12_LRHQ and an intracellular Fab | | Descriptor: | Atypical chemokine receptor 3, CID24 Fab heavy chain, CID24 Fab light chain, ... | | Authors: | Yen, Y.C, Schafer, C.T, Gustavsson, M, Handel, T.M, Tesmer, J.J.G. | | Deposit date: | 2021-10-19 | | Release date: | 2022-07-27 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structures of atypical chemokine receptor 3 reveal the basis for its promiscuity and signaling bias.
Sci Adv, 8, 2022
|
|
7SK9
 
 | | Cryo-EM structure of human ACKR3 in complex with a small molecule partial agonist CCX662, and an intracellular Fab | | Descriptor: | (1R)-4-[7-(3-carboxypropoxy)-6-methylquinolin-8-yl]-1-{[2-(4-hydroxypiperidin-1-yl)-1,3-thiazol-4-yl]methyl}-1,4-diazepan-1-ium, Atypical chemokine receptor 3, CHOLESTEROL, ... | | Authors: | Yen, Y.C, Schafer, C.T, Gustavsson, M, Handel, T.M, Tesmer, J.J.G. | | Deposit date: | 2021-10-19 | | Release date: | 2022-07-27 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structures of atypical chemokine receptor 3 reveal the basis for its promiscuity and signaling bias.
Sci Adv, 8, 2022
|
|
7SK8
 
 | | Cryo-EM structure of human ACKR3 in complex with CXCL12, a small molecule partial agonist CCX662, an extracellular Fab, and an intracellular Fab | | Descriptor: | (1R)-4-[7-(3-carboxypropoxy)-6-methylquinolin-8-yl]-1-{[2-(4-hydroxypiperidin-1-yl)-1,3-thiazol-4-yl]methyl}-1,4-diazepan-1-ium, Atypical chemokine receptor 3, CHOLESTEROL, ... | | Authors: | Yen, Y.C, Schafer, C.T, Gustavsson, M, Handel, T.M, Tesmer, J.J.G. | | Deposit date: | 2021-10-19 | | Release date: | 2022-07-27 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structures of atypical chemokine receptor 3 reveal the basis for its promiscuity and signaling bias.
Sci Adv, 8, 2022
|
|
7SK3
 
 | | Cryo-EM structure of ACKR3 in complex with CXCL12, an intracellular Fab, and an extracellular Fab | | Descriptor: | Atypical chemokine receptor 3, CHOLESTEROL, CID24 Fab heavy chain, ... | | Authors: | Yen, Y.C, Schafer, C.T, Gustavsson, M, Handel, T.M, Tesmer, J.J.G. | | Deposit date: | 2021-10-19 | | Release date: | 2022-07-27 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structures of atypical chemokine receptor 3 reveal the basis for its promiscuity and signaling bias.
Sci Adv, 8, 2022
|
|
4PKX
 
 | | The structure of a conserved Piezo channel domain reveals a novel beta sandwich fold | | Descriptor: | Protein C10C5.1, isoform i | | Authors: | Kamajaya, A, Kaiser, J, Lee, J, Reid, M, Rees, D.C. | | Deposit date: | 2014-05-15 | | Release date: | 2014-10-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | The Structure of a Conserved Piezo Channel Domain Reveals a Topologically Distinct beta Sandwich Fold.
Structure, 22, 2014
|
|
4PMQ
 
 | | Crystal structure of the Mycobacterium tuberculosis Tat-secreted protein Rv2525c in complex with L-tartrate (orthorhombic crystal form) | | Descriptor: | GLYCEROL, L(+)-TARTARIC ACID, Tat-secreted protein Rv2525c | | Authors: | Bellinzoni, M, Haouz, A, Shepard, W, Alzari, P.M. | | Deposit date: | 2014-05-22 | | Release date: | 2014-10-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Structural studies suggest a peptidoglycan hydrolase function for the Mycobacterium tuberculosis Tat-secreted protein Rv2525c.
J.Struct.Biol., 188, 2014
|
|
4PDT
 
 | | Japanese Marasmius oreades lectin | | Descriptor: | Mannose recognizing lectin, SULFATE ION | | Authors: | Noma, Y, Shimokawa, M, Maeganeku, C, Motoshima, H, Watanabe, K, Minami, Y, Yagi, F. | | Deposit date: | 2014-04-22 | | Release date: | 2015-04-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The structure of Japanese Marasmius oreades lectin at 1.40 Angstroms resolution.
To Be Published
|
|
4PSX
 
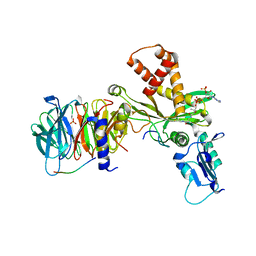 | | Crystal structure of histone acetyltransferase complex | | Descriptor: | COENZYME A, Histone H3, Histone H4, ... | | Authors: | Yang, M, Li, Y. | | Deposit date: | 2014-03-08 | | Release date: | 2014-07-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.509 Å) | | Cite: | Hat2p recognizes the histone H3 tail to specify the acetylation of the newly synthesized H3/H4 heterodimer by the Hat1p/Hat2p complex
Genes Dev., 28, 2014
|
|
4P8Z
 
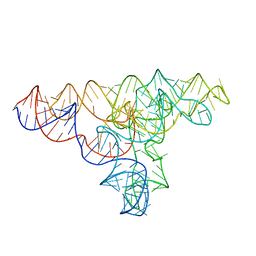 | | Speciation of a group I intron into a lariat capping ribozyme | | Descriptor: | Didymium iridis partial IGS, 18S rRNA gene, I-DirI gene and partial ITS1 | | Authors: | Meyer, M, Nielsen, H, Olieric, V, Roblin, P, Johansen, S.D, Westhof, E, Masquida, B. | | Deposit date: | 2014-04-01 | | Release date: | 2014-05-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.85 Å) | | Cite: | Speciation of a group I intron into a lariat capping ribozyme.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4PCF
 
 | |
