7FAS
 
 | | VAR2CSA 3D7 ectodomain core region | | Descriptor: | Erythrocyte membrane protein 1, PfEMP1 | | Authors: | Wang, L, Zhaoning, W. | | Deposit date: | 2021-07-07 | | Release date: | 2021-11-17 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | The molecular mechanism of cytoadherence to placental or tumor cells through VAR2CSA from Plasmodium falciparum.
Cell Discov, 7, 2021
|
|
4Y4R
 
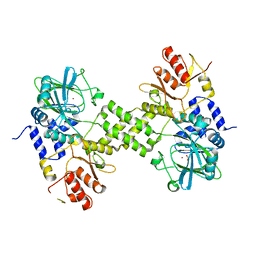 | | Crystal structure of ribosomal oxygenase NO66 dimer mutant | | Descriptor: | ACETATE ION, Bifunctional lysine-specific demethylase and histidyl-hydroxylase NO66, NICKEL (II) ION | | Authors: | Wang, C, Hang, T, Zang, J. | | Deposit date: | 2015-02-11 | | Release date: | 2015-10-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure of the JmjC domain-containing protein NO66 complexed with ribosomal protein Rpl8.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
6W3I
 
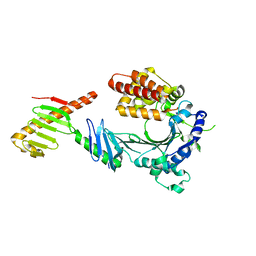 | | Crystal structure of a FAM46C mutant in complex with Plk4 | | Descriptor: | Serine/threonine-protein kinase PLK4, Terminal nucleotidyltransferase 5C | | Authors: | Chen, H, Shang, G.J, Lu, D.F, Zhang, X.W. | | Deposit date: | 2020-03-09 | | Release date: | 2020-05-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.802 Å) | | Cite: | Structural and Functional Analyses of the FAM46C/Plk4 Complex.
Structure, 28, 2020
|
|
3K3O
 
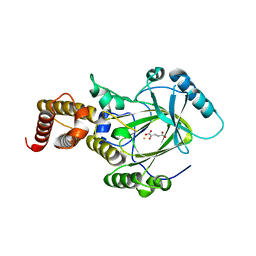 | | Crystal structure of the catalytic core domain of human PHF8 complexed with alpha-ketoglutarate | | Descriptor: | 2-OXOGLUTARIC ACID, FE (II) ION, PHD finger protein 8 | | Authors: | Yu, L, Wang, Y, Huang, S, Wang, J, Deng, Z, Wu, W, Gong, W, Chen, Z. | | Deposit date: | 2009-10-03 | | Release date: | 2010-01-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural insights into a novel histone demethylase PHF8
Cell Res., 20, 2010
|
|
4YR6
 
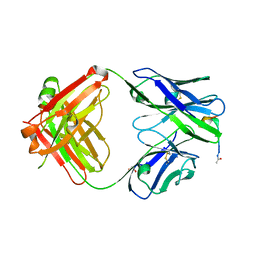 | | Fab fragment of 5G6 in complex with epitope peptide | | Descriptor: | ACE-LYS-LEU-ARG-GLY-VAL-LEU-GLN-GLY-HIS-LEU, GLYCEROL, heavy chain of 5G6, ... | | Authors: | Tao, Y, Mo, X, Li, R. | | Deposit date: | 2015-03-14 | | Release date: | 2016-03-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Structural basis for the specific inhibition of glycoprotein Ib alpha shedding by an inhibitory antibody.
Sci Rep, 6, 2016
|
|
3K3N
 
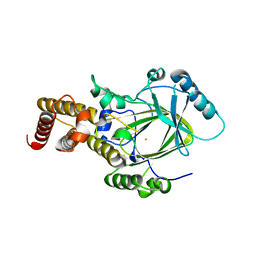 | | Crystal structure of the catalytic core domain of human PHF8 | | Descriptor: | FE (II) ION, PHD finger protein 8 | | Authors: | Yu, L, Wang, Y, Huang, S, Wang, J, Deng, Z, Wu, W, Gong, W, Chen, Z. | | Deposit date: | 2009-10-03 | | Release date: | 2010-01-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural insights into a novel histone demethylase PHF8
Cell Res., 20, 2010
|
|
6W36
 
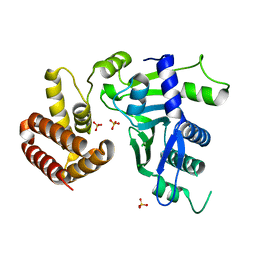 | | Crystal structure of FAM46C | | Descriptor: | SULFATE ION, Terminal nucleotidyltransferase 5C | | Authors: | Shang, G.J, Zhang, X.W, Chen, H, Lu, D.F. | | Deposit date: | 2020-03-09 | | Release date: | 2020-05-06 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.854 Å) | | Cite: | Structural and Functional Analyses of the FAM46C/Plk4 Complex.
Structure, 28, 2020
|
|
7Y0Y
 
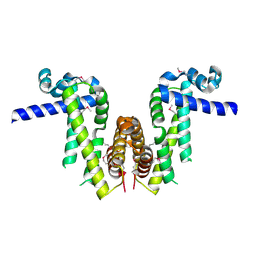 | | Crystal structure of Pseudomonas aeruginosa PvrA (SeMet) | | Descriptor: | TetR family transcriptional regulator | | Authors: | Liang, H, Zhang, Q, Bartlam, M. | | Deposit date: | 2022-06-06 | | Release date: | 2023-02-08 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Regulatory and structural mechanisms of PvrA-mediated regulation of the PQS quorum-sensing system and PHA biosynthesis in Pseudomonas aeruginosa.
Nucleic Acids Res., 51, 2023
|
|
6W38
 
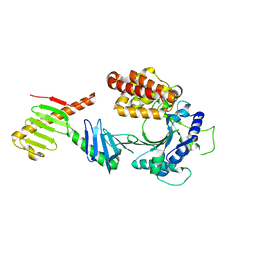 | | Crystal structure of the FAM46C/Plk4 complex | | Descriptor: | Serine/threonine-protein kinase PLK4, Terminal nucleotidyltransferase 5C | | Authors: | Chen, H, Lu, D.F, Shang, G.J, Zhang, X.W. | | Deposit date: | 2020-03-09 | | Release date: | 2020-05-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (4.48 Å) | | Cite: | Structural and Functional Analyses of the FAM46C/Plk4 Complex.
Structure, 28, 2020
|
|
6W3J
 
 | | Crystal structure of the FAM46C/Plk4/Cep192 complex | | Descriptor: | Centrosomal protein of 192 kDa, Serine/threonine-protein kinase PLK4, Terminal nucleotidyltransferase 5C | | Authors: | Chen, H, Lu, D.F, Shang, G.J, Zhang, X.W. | | Deposit date: | 2020-03-09 | | Release date: | 2020-05-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (4.385 Å) | | Cite: | Structural and Functional Analyses of the FAM46C/Plk4 Complex.
Structure, 28, 2020
|
|
7XA7
 
 | | Crystal structure of SARS-CoV-2 receptor-binding domain in complex with intermediate horseshoe bat ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme, Spike protein S1, ... | | Authors: | Tang, L.F, Zhang, D, Han, P, Qi, J.X. | | Deposit date: | 2022-03-17 | | Release date: | 2022-12-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.31 Å) | | Cite: | Structural basis of SARS-CoV-2 and its variants binding to intermediate horseshoe bat ACE2.
Int J Biol Sci, 18, 2022
|
|
1YF3
 
 | | T4Dam in Complex with AdoHcy and 13-mer Oligonucleotide Making Non- and Semi-specific (~1/4) Contact | | Descriptor: | 5'-D(*AP*CP*CP*AP*TP*GP*AP*TP*CP*TP*GP*AP*C)-3', 5'-D(*TP*GP*TP*CP*AP*GP*AP*TP*CP*AP*TP*GP*G)-3', DNA adenine methylase, ... | | Authors: | Horton, J.R, Liebert, K, Hattman, S, Jeltsch, A, Cheng, X. | | Deposit date: | 2004-12-30 | | Release date: | 2005-05-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Transition from Nonspecific to Specific DNA Interactions along the Substrate-Recognition Pathway of Dam Methyltransferase.
Cell(Cambridge,Mass.), 121, 2005
|
|
1YFL
 
 | | T4Dam in Complex with Sinefungin and 16-mer Oligonucleotide Showing Semi-specific and Specific Contact and Flipped Base | | Descriptor: | 5'-D(P*TP*CP*AP*CP*AP*GP*GP*AP*TP*CP*CP*TP*GP*TP*GP*A)-3', DNA adenine methylase, SINEFUNGIN | | Authors: | Horton, J.R, Liebert, K, Hattman, S, Jeltsch, A, Cheng, X. | | Deposit date: | 2005-01-03 | | Release date: | 2005-05-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | Transition from Nonspecific to Specific DNA Interactions along the Substrate-Recognition Pathway of Dam Methyltransferase.
Cell(Cambridge,Mass.), 121, 2005
|
|
3MTS
 
 | | Chromo Domain of Human Histone-Lysine N-Methyltransferase SUV39H1 | | Descriptor: | Histone-lysine N-methyltransferase SUV39H1 | | Authors: | Lam, R, Li, Z, Wang, J, Crombet, L, Walker, J.R, Ouyang, H, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-04-30 | | Release date: | 2010-06-30 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of the Human SUV39H1 Chromodomain and Its Recognition of Histone H3K9me2/3.
Plos One, 7, 2012
|
|
7V6Y
 
 | | Cryo-EM structure of Patched in lipid nanodisc - the wildtype, 3.5 angstrom (re-processed with dataset of 7dzq) | | Descriptor: | (2S)-2-azanyl-3-[[(2S)-3-butanoyloxy-2-dec-9-enoyloxy-propoxy]-oxidanyl-phosphoryl]oxy-propanoic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Luo, Y, Zhao, Y, Qu, Q, Li, D. | | Deposit date: | 2021-08-20 | | Release date: | 2021-09-22 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM study of patched in lipid nanodisc suggests a structural basis for its clustering in caveolae.
Structure, 29, 2021
|
|
7V6Z
 
 | | Cryo-EM structure of Patched1 (V1084A mutant) in lipid nanodisc, 3.64 angstrom (reprocessed with the dataset of 7dzp) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, ... | | Authors: | Luo, Y, Zhao, Y, Qu, Q, Li, D. | | Deposit date: | 2021-08-20 | | Release date: | 2021-09-22 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (3.64 Å) | | Cite: | Cryo-EM study of patched in lipid nanodisc suggests a structural basis for its clustering in caveolae.
Structure, 29, 2021
|
|
1YFJ
 
 | | T4Dam in Complex with AdoHcy and 15-mer Oligonucleotide Showing Semi-specific and Specific Contact | | Descriptor: | 5'-D(*TP*CP*AP*CP*AP*GP*GP*AP*TP*CP*CP*TP*GP*TP*G)-3', CALCIUM ION, CHLORIDE ION, ... | | Authors: | Horton, J.R, Liebert, K, Hattman, S, Jeltsch, A, Cheng, X. | | Deposit date: | 2005-01-02 | | Release date: | 2005-05-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Transition from Nonspecific to Specific DNA Interactions along the Substrate-Recognition Pathway of Dam Methyltransferase.
Cell(Cambridge,Mass.), 121, 2005
|
|
7Y0Z
 
 | | Crystal structure of Pseudomonas aeruginosa PvrA | | Descriptor: | TetR family transcriptional regulator | | Authors: | Liang, H, Zhang, Q, Bartlam, M. | | Deposit date: | 2022-06-06 | | Release date: | 2023-02-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Regulatory and structural mechanisms of PvrA-mediated regulation of the PQS quorum-sensing system and PHA biosynthesis in Pseudomonas aeruginosa.
Nucleic Acids Res., 51, 2023
|
|
2LI6
 
 | |
4LA2
 
 | | Crystal structure of dimethylsulphoniopropionate (DMSP) lyase DddQ | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Dimethylsulphoniopropionate (DMSP) lyase DddQ, ZINC ION | | Authors: | Zhang, Y, Li, C. | | Deposit date: | 2013-06-19 | | Release date: | 2014-01-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Molecular insight into bacterial cleavage of oceanic dimethylsulfoniopropionate into dimethyl sulfide
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4LX4
 
 | | Crystal Structure Determination of Pseudomonas stutzeri endoglucanase Cel5A using a Twinned Data Set | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Endoglucanase(Endo-1,4-beta-glucanase)protein | | Authors: | Dutoit, R, Delsaute, M, Berlemont, R, Van Elder, D, Galleni, M, Bauvois, C. | | Deposit date: | 2013-07-29 | | Release date: | 2014-07-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.556 Å) | | Cite: | Crystal structure determination of Pseudomonas stutzeri A1501 endoglucanase Cel5A: the search for a molecular basis for glycosynthesis in GH5_5 enzymes.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
8D4L
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) S144A Mutant | | Descriptor: | 3C-like proteinase nsp5 | | Authors: | Lewandowski, E.M, Hu, Y, Tan, H, Wang, J, Chen, Y. | | Deposit date: | 2022-06-02 | | Release date: | 2022-07-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Naturally occurring mutations of SARS-CoV-2 main protease confer drug resistance to nirmatrelvir.
Biorxiv, 2022
|
|
8SEH
 
 | | PHF Tau from Down Syndrome | | Descriptor: | Microtubule-associated protein tau | | Authors: | Hoq, M.R, Bharath, S.R, Jiang, W, Vago, F.S. | | Deposit date: | 2023-04-10 | | Release date: | 2024-04-03 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Cryo-EM structures of amyloid-beta and tau filaments in Down syndrome.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8SEK
 
 | |
8SEL
 
 | |
