4WH1
 
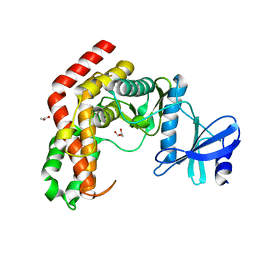 | | N-Acetylhexosamine 1-kinase (ligand free) | | Descriptor: | ACETIC ACID, GLYCEROL, N-acetylhexosamine 1-kinase | | Authors: | Sato, M, Arakawa, T, Nam, Y.W, Nishimoto, M, Kitaoka, M, Fushinobu, S. | | Deposit date: | 2014-09-19 | | Release date: | 2015-02-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Open-close structural change upon ligand binding and two magnesium ions required for the catalysis of N-acetylhexosamine 1-kinase
Biochim.Biophys.Acta, 1854, 2015
|
|
4WH3
 
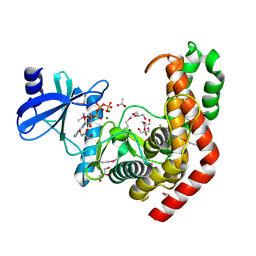 | | N-acetylhexosamine 1-kinase in complex with ATP | | Descriptor: | ACETIC ACID, ADENOSINE-5'-TRIPHOSPHATE, GLYCEROL, ... | | Authors: | Sato, M, Arakawa, T, Nam, Y.W, Nishimoto, M, Kitaoka, M, Fushinobu, S. | | Deposit date: | 2014-09-19 | | Release date: | 2015-02-18 | | Last modified: | 2020-02-05 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Open-close structural change upon ligand binding and two magnesium ions required for the catalysis of N-acetylhexosamine 1-kinase
Biochim.Biophys.Acta, 1854, 2015
|
|
8CXC
 
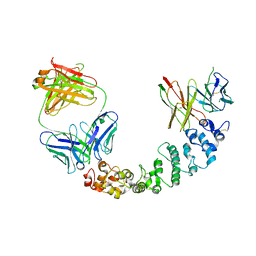 | | Novel Anti-Mesothelin Antibodies Enable Crystallography of the Intact Mesothelin Ectodo- main and Engineering of Potent, T cell-engaging Bispecific Therapeutics | | Descriptor: | 3F2 Antibody heavy chain, 3F2 Antibody light chain, Mesothelin, ... | | Authors: | Bandaranayake, A.D, Rupert, P.B, Lin, I, Pilat, K, Ruff, R.O, Friend, D.J, Chan, M.K, Clarke, M, Carter, J, Meshinchi, S, Mehlin, C, Olson, J.M, Strong, R.K, Correnti, C.E. | | Deposit date: | 2022-05-20 | | Release date: | 2023-06-07 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (4.31 Å) | | Cite: | Novel mesothelin antibodies enable crystallography of the intact mesothelin ectodomain and engineering of potent, T cell-engaging bispecific therapeutics
Front. Drug Discov., 3, 2023
|
|
8CYH
 
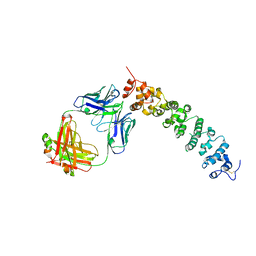 | | Novel Anti-Mesothelin Antibodies Enable Crystallography of the Intact Mesothelin Ectodo- main and Engineering of Potent, T cell-engaging Bispecific Therapeutics | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, A12 antibody heavy chain, A12 antibody light chain, ... | | Authors: | Bandaranayake, A.D, Rupert, P.B, Lin, I, Pilat, K, Ruff, R.O, Friend, D.J, Chan, M.K, Clarke, M, Carter, J, Meshinchi, S, Mehlin, C, Olson, J.M, Strong, R.K, Correnti, C.E. | | Deposit date: | 2022-05-23 | | Release date: | 2023-06-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.38 Å) | | Cite: | Novel mesothelin antibodies enable crystallography of the intact mesothelin ectodomain and engineering of potent, T cell-engaging bispecific therapeutics
Front. Drug Discov., 3, 2023
|
|
8CZ8
 
 | | Novel Anti-Mesothelin Antibodies Enable Crystallography of the Intact Mesothelin Ectodo- main and Engineering of Potent, T cell-engaging Bispecific Therapeutics | | Descriptor: | Mesothelin, cleaved form, SULFATE ION, ... | | Authors: | Bandaranayake, A.D, Rupert, P.B, Lin, I, Pilat, K, Ruff, R.O, Friend, D.J, Chan, M.K, Clarke, M, Carter, J, Meshinchi, S, Mehlin, C, Olson, J.M, Strong, R.K, Correnti, C.E. | | Deposit date: | 2022-05-24 | | Release date: | 2023-06-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Novel mesothelin antibodies enable crystallography of the intact mesothelin ectodomain and engineering of potent, T cell-engaging bispecific therapeutics
Front. Drug Discov., 3, 2023
|
|
6K8Q
 
 | | Solution structure of the intermembrane space domain of the mitochondrial import protein Tim21 from S. cerevisiae | | Descriptor: | Mitochondrial import inner membrane translocase subunit TIM21 | | Authors: | Bala, S, Shinya, S, Srivastava, A, Shimada, A, Kobayashi, N, Kojima, C, Tama, F, Miyashita, O, Kohda, D. | | Deposit date: | 2019-06-13 | | Release date: | 2019-09-11 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Crystal contact-free conformation of an intrinsically flexible loop in protein crystal: Tim21 as the case study.
Biochim Biophys Acta Gen Subj, 1864, 2020
|
|
5DGR
 
 | | Crystal structure of GH9 exo-beta-D-glucosaminidase PBPRA0520, glucosamine complex | | Descriptor: | 2-amino-2-deoxy-beta-D-glucopyranose, Putative endoglucanase-related protein, SODIUM ION | | Authors: | Suzuki, K, Honda, Y, Fushinobu, S. | | Deposit date: | 2015-08-28 | | Release date: | 2015-12-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The crystal structure of an inverting glycoside hydrolase family 9 exo-beta-D-glucosaminidase and the design of glycosynthase.
Biochem.J., 473, 2016
|
|
5BXR
 
 | | LNBase in complex with LNB-NHAcDNJ | | Descriptor: | 2-ACETAMIDO-1,2-DIDEOXYNOJIRMYCIN, Lacto-N-biosidase, SULFATE ION, ... | | Authors: | Ito, T, Arakawa, T, Fushinobu, S. | | Deposit date: | 2015-06-09 | | Release date: | 2015-09-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Gaining insight into the catalysis by GH20 lacto-N-biosidase using small molecule inhibitors and structural analysis
Chem.Commun.(Camb.), 51, 2015
|
|
5BXT
 
 | | LNBase in complex with LNB-NHAcAUS | | Descriptor: | Lacto-N-biosidase, N-{[(1R,2R,3R,7S,7aR)-1,2,7-trihydroxyhexahydro-1H-pyrrolizin-3-yl]methyl}acetamide, SULFATE ION, ... | | Authors: | Ito, T, Arakawa, T, Fushinobu, S. | | Deposit date: | 2015-06-09 | | Release date: | 2015-09-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Gaining insight into the catalysis by GH20 lacto-N-biosidase using small molecule inhibitors and structural analysis
Chem.Commun.(Camb.), 51, 2015
|
|
5BXP
 
 | | LNBase in complex with LNB-LOGNAc | | Descriptor: | Lacto-N-biosidase, SULFATE ION, beta-D-galactopyranose-(1-3)-N-acetylglucosaminono-1,5-lactone (Z)-oxime | | Authors: | Ito, T, Arakawa, T, Fushinobu, S. | | Deposit date: | 2015-06-09 | | Release date: | 2015-09-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Gaining insight into the catalysis by GH20 lacto-N-biosidase using small molecule inhibitors and structural analysis
Chem.Commun.(Camb.), 51, 2015
|
|
5GZK
 
 | | Endo-beta-1,2-glucanase from Chitinophaga pinensis - sophorotriose and glucose complex | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, PENTAETHYLENE GLYCOL, ... | | Authors: | Abe, K, Nakajima, M, Arakawa, T, Fushinobu, S, Taguchi, H. | | Deposit date: | 2016-09-28 | | Release date: | 2017-03-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Biochemical and structural analyses of a bacterial endo-beta-1,2-glucanase reveal a new glycoside hydrolase family
J. Biol. Chem., 292, 2017
|
|
5BXS
 
 | | LNBase in complex with LNB-NHAcCAS | | Descriptor: | 6-ACETAMIDO-6-DEOXY-CASTANOSPERMINE, Lacto-N-biosidase, SULFATE ION, ... | | Authors: | Ito, T, Arakawa, T, Fushinobu, S. | | Deposit date: | 2015-06-09 | | Release date: | 2015-09-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Gaining insight into the catalysis by GH20 lacto-N-biosidase using small molecule inhibitors and structural analysis
Chem.Commun.(Camb.), 51, 2015
|
|
5GZH
 
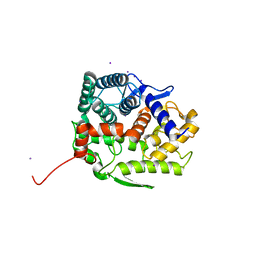 | | Endo-beta-1,2-glucanase from Chitinophaga pinensis - ligand free form | | Descriptor: | Endo-beta-1,2-glucanase, IODIDE ION, alpha-D-glucopyranose-(1-1)-alpha-D-glucopyranose | | Authors: | Abe, K, Nakajima, M, Arakawa, T, Fushinobu, S, Taguchi, H. | | Deposit date: | 2016-09-28 | | Release date: | 2017-03-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Biochemical and structural analyses of a bacterial endo-beta-1,2-glucanase reveal a new glycoside hydrolase family
J. Biol. Chem., 292, 2017
|
|
1ITP
 
 | | Solution Structure of POIA1 | | Descriptor: | proteinase A inhibitor 1 | | Authors: | Sasakawa, H, Yoshinaga, S, Kojima, S, Tamura, A. | | Deposit date: | 2002-01-23 | | Release date: | 2002-02-13 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structure of POIA1, a homologous protein to the propeptide of subtilisin: implication for protein foldability and the function as an intramolecular chaperone.
J.Mol.Biol., 317, 2002
|
|
8YK2
 
 | | Blood group B alpha-1,3-galactosidase AgaBb from Bifidobacterium bifidum, construct T7-tag_24-700 | | Descriptor: | Alpha-galactosidase, GLYCEROL, SODIUM ION, ... | | Authors: | Kashima, T, Akama, M, Ashida, H, Fushinobu, S. | | Deposit date: | 2024-03-04 | | Release date: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Crystal Structure of Bifidobacterium bifidum Glycoside Hydrolase Family 110 alpha-Galactosidase Specific for Blood Group B Antigen
J.Appl.Glyosci., 2024
|
|
8WUC
 
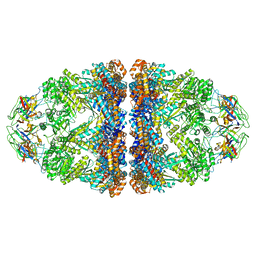 | | Cryo-EM structure of H. thermoluteolus GroEL-GroES2 football complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Chaperonin GroEL, Co-chaperonin GroES, ... | | Authors: | Liao, Z, Gopalasingam, C.C, Kameya, M, Gerle, C, Shigematsu, H, Ishii, M, Arakawa, T, Fushinobu, S. | | Deposit date: | 2023-10-20 | | Release date: | 2024-03-27 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Structural insights into thermophilic chaperonin complexes.
Structure, 32, 2024
|
|
6JI2
 
 | | Crystal structure of archaeal ribosomal protein aP1, aPelota, and GTP-bound aEF1A complex | | Descriptor: | Archaeal ribosomal stalk protein aP1, Elongation factor 1-alpha, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Maruyama, K, Imai, H, Kawamura, M, Ishino, S, Ishino, Y, Ito, K, Uchiumi, T. | | Deposit date: | 2019-02-20 | | Release date: | 2019-11-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Switch of the interactions between the ribosomal stalk and EF1A in the GTP- and GDP-bound conformations.
Sci Rep, 9, 2019
|
|
8WUW
 
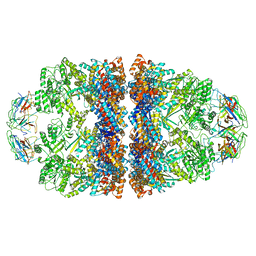 | | Cryo-EM structure of H. thermophilus GroEL-GroES2 asymmetric football complex | | Descriptor: | Chaperonin GroEL, Co-chaperonin GroES, MAGNESIUM ION, ... | | Authors: | Liao, Z, Gopalasingam, C.C, Kameya, M, Gerle, C, Shigematsu, H, Ishii, M, Arakawa, T, Fushinobu, S. | | Deposit date: | 2023-10-21 | | Release date: | 2024-03-27 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural insights into thermophilic chaperonin complexes.
Structure, 32, 2024
|
|
8YK3
 
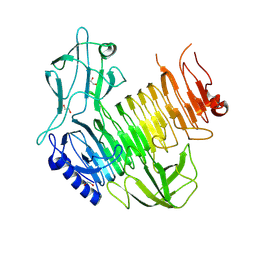 | | Blood group B alpha-1,3-galactosidase AgaBb from Bifidobacterium bifidum, construct T7-tag_24-673 | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Alpha-galactosidase, ... | | Authors: | Kashima, T, Ashida, H, Fushinobu, S. | | Deposit date: | 2024-03-04 | | Release date: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystal Structure of Bifidobacterium bifidum Glycoside Hydrolase Family 110 alpha-Galactosidase Specific for Blood Group B Antigen
J.Appl.Glyosci., 2024
|
|
8WUX
 
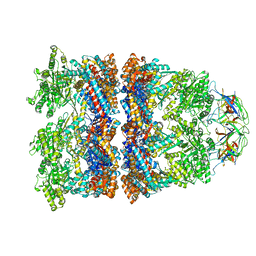 | | Cryo-EM structure of H. thermophilus GroEL-GroES bullet complex | | Descriptor: | Chaperonin GroEL, Co-chaperonin GroES, MAGNESIUM ION, ... | | Authors: | Liao, Z, Gopalasingam, C.C, Kameya, M, Gerle, C, Shigematsu, H, Ishii, M, Arakawa, T, Fushinobu, S. | | Deposit date: | 2023-10-21 | | Release date: | 2024-03-27 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural insights into thermophilic chaperonin complexes.
Structure, 32, 2024
|
|
8WU4
 
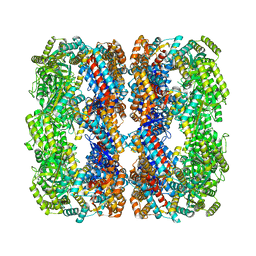 | | Cryo-EM structure of native H. thermoluteolus TH-1 GroEL | | Descriptor: | Chaperonin GroEL | | Authors: | Liao, Z, Gopalasingam, C.C, Kameya, M, Gerle, C, Shigematsu, H, Ishii, M, Arakawa, T, Fushinobu, S. | | Deposit date: | 2023-10-20 | | Release date: | 2024-03-27 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural insights into thermophilic chaperonin complexes.
Structure, 32, 2024
|
|
8YK1
 
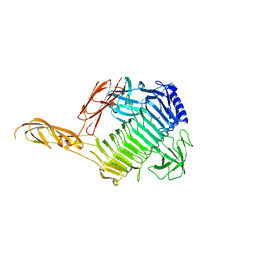 | |
2D0D
 
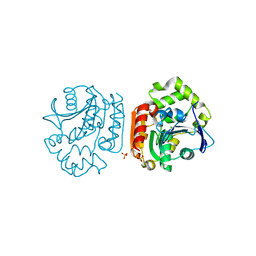 | | Crystal Structure of a Meta-cleavage Product Hydrolase (CumD) A129V Mutant | | Descriptor: | 2-hydroxy-6-oxo-7-methylocta-2,4-dienoate hydrolase, CHLORIDE ION, PHOSPHATE ION | | Authors: | Jun, S.Y, Fushinobu, S, Nojiri, H, Omori, T, Shoun, H, Wakagi, T. | | Deposit date: | 2005-08-01 | | Release date: | 2006-06-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Improving the catalytic efficiency of a meta-cleavage product hydrolase (CumD) from Pseudomonas fluorescens IP01
Biochim.Biophys.Acta, 1764, 2006
|
|
3ACT
 
 | |
3AHG
 
 | | H64A mutant of Phosphoketolase from Bifidobacterium Breve complexed with a tricyclic ring form of thiamine diphosphate | | Descriptor: | 1,2-ETHANEDIOL, 2-[(9aR)-2,7-dimethyl-9a,10-dihydro-5H-pyrimido[4,5-d][1,3]thiazolo[3,2-a]pyrimidin-8-yl]ethyl trihydrogen diphosphate, MAGNESIUM ION, ... | | Authors: | Suzuki, R, Katayama, T, Kim, B.-J, Wakagi, T, Shoun, H, Ashida, H, Yamamoto, K, Fushinobu, S. | | Deposit date: | 2010-04-22 | | Release date: | 2010-08-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structures of phosphoketolase: thiamine diphosphate-dependent dehydration mechanism
J.Biol.Chem., 285, 2010
|
|
