7AEN
 
 | | Galectin-8 N-terminal carbohydrate recognition domain in complex with methyl 3-O-((7-carboxy)quinolin-2-yl)-methoxy)-beta-D-galactopyranoside | | Descriptor: | CHLORIDE ION, GLYCEROL, Isoform 2 of Galectin-8, ... | | Authors: | Hassan, M, Klavern, V.S, Hakansson, M, Anderluh, M, Tomasic, T, Jakopin, Z, Nilsson, J.U, Kovacic, R, Walse, B, Diehl, C. | | Deposit date: | 2020-09-17 | | Release date: | 2021-07-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure-Guided Design of d-Galactal Derivatives with High Affinity and Selectivity for the Galectin-8 N-Terminal Domain
Acs Med.Chem.Lett., 12, 2021
|
|
6SNL
 
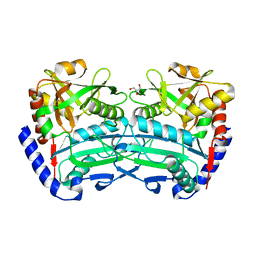 | | (R)-selective amine transaminase from Exophiala sideris | | Descriptor: | CHLORIDE ION, GLYCEROL, HEXAETHYLENE GLYCOL, ... | | Authors: | Telzerow, A, Hakansson, M, Steiner, K. | | Deposit date: | 2019-08-26 | | Release date: | 2020-12-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.129 Å) | | Cite: | Expanding the Toolbox of R-Selective Amine Transaminases by Identification and Characterization of New Members.
Chembiochem, 22, 2021
|
|
7S0J
 
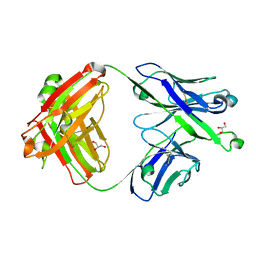 | | Crystal structure of Epstein-Barr virus gH/gL targeting antibody 769B10 | | Descriptor: | 769B10 Fab Heavy chain, 769B10 Fab Light chain, GLYCEROL | | Authors: | Chen, W.-H, Kanekiyo, M, Cohen, J.I, Joyce, M.G. | | Deposit date: | 2021-08-30 | | Release date: | 2022-11-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Epstein-Barr virus gH/gL has multiple sites of vulnerability for virus neutralization and fusion inhibition.
Immunity, 55, 2022
|
|
7S1B
 
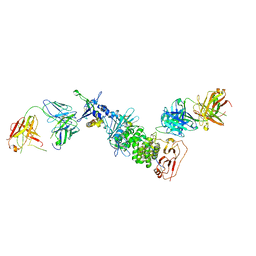 | | Crystal structure of Epstein-Barr virus glycoproteins gH/gL/gp42-peptide in complex with human neutralizing antibodies 769C2 and 770F7 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 769C2 Fab heavy chain, 769C2 Fab light chain, ... | | Authors: | Chen, W.-H, Cohen, J.I, Kanekiyo, M, Joyce, M.G. | | Deposit date: | 2021-09-02 | | Release date: | 2022-11-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.03 Å) | | Cite: | Epstein-Barr virus gH/gL has multiple sites of vulnerability for virus neutralization and fusion inhibition.
Immunity, 55, 2022
|
|
7S07
 
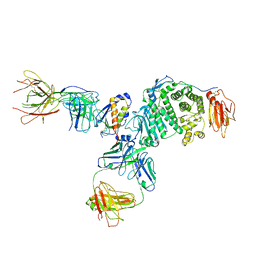 | | Crystal structure of Epstein-Barr virus glycoprotein gH/gL/gp42-peptide in complex with human neutralizing antibodies 769B10 and 769C2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 769B10 Fab heavy chain, 769B10 Fab light chain, ... | | Authors: | Chen, W.-H, Kanekiyo, M, Cohen, J.I, Joyce, M.G. | | Deposit date: | 2021-08-30 | | Release date: | 2022-11-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.29 Å) | | Cite: | Epstein-Barr virus gH/gL has multiple sites of vulnerability for virus neutralization and fusion inhibition.
Immunity, 55, 2022
|
|
8R5J
 
 | | Crystal structure of MERS-CoV main protease | | Descriptor: | Non-structural protein 11 | | Authors: | Balcomb, B.H, Fairhead, M, Koekemoer, L, Lithgo, R.M, Aschenbrenner, J.C, Chandran, A.V, Godoy, A.S, Lukacik, P, Marples, P.G, Mazzorana, M, Ni, X, Strain-Damerell, C, Thompson, W, Tomlinson, C.W.E, Wild, C, Winokan, M, Fearon, D, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-11-16 | | Release date: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.898 Å) | | Cite: | Crystal structure of MERS-CoV main protease
To Be Published
|
|
4XSJ
 
 | | Crystal structure of the N-terminal domain of the human mitochondrial calcium uniporter fused with T4 lysozyme | | Descriptor: | Lysozyme,Calcium uniporter protein, mitochondrial, SULFATE ION | | Authors: | Lee, Y, Min, C.K, Kim, T.G, Song, H.K, Lim, Y, Kim, D, Shin, K, Kang, M, Kang, J.Y, Youn, H.-S, Lee, J.-G, An, J.Y, Park, K.R, Lim, J.J, Kim, J.H, Kim, J.H, Park, Z.Y, Kim, Y.-S, Wang, J, Kim, D.H, Eom, S.H. | | Deposit date: | 2015-01-22 | | Release date: | 2015-09-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure and function of the N-terminal domain of the human mitochondrial calcium uniporter.
Embo Rep., 16, 2015
|
|
4XTB
 
 | | Crystal structure of the N-terminal domain of the human mitochondrial calcium uniporter | | Descriptor: | Calcium uniporter protein, mitochondrial, TETRAETHYLENE GLYCOL | | Authors: | Lee, Y, Min, C.K, Kim, T.G, Song, H.K, Lim, Y, Kim, D, Shin, K, Kang, M, Kang, J.Y, Youn, H.-S, Lee, J.-G, An, J.Y, Park, K.R, Lim, J.J, Kim, J.H, Kim, J.H, Park, Z.Y, Kim, Y.-S, Wang, J, Kim, D.H, Eom, S.H. | | Deposit date: | 2015-01-23 | | Release date: | 2015-09-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure and function of the N-terminal domain of the human mitochondrial calcium uniporter.
Embo Rep., 16, 2015
|
|
6XU3
 
 | | (R)-selective amine transaminase from Shinella sp. | | Descriptor: | 3-AMINOBENZOIC ACID, CHLORIDE ION, Class IV aminotransferase, ... | | Authors: | Telzerow, A, Hakansson, M, Steiner, K. | | Deposit date: | 2020-01-17 | | Release date: | 2020-12-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Expanding the Toolbox of R-Selective Amine Transaminases by Identification and Characterization of New Members.
Chembiochem, 22, 2021
|
|
6Q4S
 
 | | Crystal structure of a-eudesmol synthase | | Descriptor: | CHLORIDE ION, Pentalenene synthase | | Authors: | Correia Cordeiro, R.S, Hakansson, M, Logan, D.T, Kourist, R. | | Deposit date: | 2018-12-06 | | Release date: | 2018-12-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Discovery of three novel sesquiterpene synthases from Streptomyces chartreusis NRRL 3882 and crystal structure of an alpha-eudesmol synthase.
J.Biotechnol., 297, 2019
|
|
6A17
 
 | | Crystal structure of CYP90B1 in complex with brassinazole | | Descriptor: | (2R,3S)-4-(4-chlorophenyl)-2-phenyl-3-(1H-1,2,4-triazol-1-yl)butan-2-ol, CHLORIDE ION, Cytochrome P450 90B1, ... | | Authors: | Fujiyama, K, Hino, T, Kanadani, M, Mizutani, M, Nagano, S. | | Deposit date: | 2018-06-06 | | Release date: | 2019-06-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Structural insights into a key step of brassinosteroid biosynthesis and its inhibition.
Nat.Plants, 5, 2019
|
|
6A15
 
 | | Structure of CYP90B1 in complex with cholesterol | | Descriptor: | CHLORIDE ION, CHOLESTEROL, Cytochrome P450 90B1, ... | | Authors: | Fujiyama, K, Hino, T, Kanadani, M, Mizutani, M, Nagano, S. | | Deposit date: | 2018-06-06 | | Release date: | 2019-06-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Structural insights into a key step of brassinosteroid biosynthesis and its inhibition.
Nat.Plants, 5, 2019
|
|
6A18
 
 | | Crystal structure of CYP90B1 in complex with 1,6-hexandiol | | Descriptor: | CHLORIDE ION, Cytochrome P450 90B1, GLYCEROL, ... | | Authors: | Fujiyama, K, Hino, T, Kanadani, M, Mizutani, M, Nagano, S. | | Deposit date: | 2018-06-06 | | Release date: | 2019-06-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Structural insights into a key step of brassinosteroid biosynthesis and its inhibition.
Nat.Plants, 5, 2019
|
|
6A16
 
 | | Crystal structure of CYP90B1 in complex with uniconazole | | Descriptor: | (1E,3S)-1-(4-chlorophenyl)-4,4-dimethyl-2-(1H-1,2,4-triazol-1-yl)pent-1-en-3-ol, CHLORIDE ION, Cytochrome P450 90B1, ... | | Authors: | Fujiyama, K, Hino, T, Kanadani, M, Mizutani, M, Nagano, S. | | Deposit date: | 2018-06-06 | | Release date: | 2019-06-12 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.998 Å) | | Cite: | Structural insights into a key step of brassinosteroid biosynthesis and its inhibition.
Nat.Plants, 5, 2019
|
|
8P8H
 
 | | Crystal structure of HHD2 domain of hRTEL1 | | Descriptor: | Regulator of telomere elongation helicase 1 | | Authors: | Hegde, R.P, Kanade, M, Cortone, G, Graewert, M, Longo, A, Gonzalez, A, Chaves-Arquero, B, Blanco, F.J, Napolitano, L.M.R, Onesti, S. | | Deposit date: | 2023-06-01 | | Release date: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of HHD2 domain of hRTEL1
To Be Published
|
|
5WVO
 
 | | Crystal structure of DNMT1 RFTS domain in complex with K18/K23 mono-ubiquitylated histone H3 | | Descriptor: | DNA (cytosine-5)-methyltransferase 1, Histone H3.1, Ubiquitin, ... | | Authors: | Ishiyama, S, Nishiyama, A, Nakanishi, M, Arita, K. | | Deposit date: | 2016-12-28 | | Release date: | 2017-11-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.997 Å) | | Cite: | Structure of the Dnmt1 Reader Module Complexed with a Unique Two-Mono-Ubiquitin Mark on Histone H3 Reveals the Basis for DNA Methylation Maintenance
Mol. Cell, 68, 2017
|
|
3WWK
 
 | | Crystal structure of CLEC-2 in complex with rhodocytin | | Descriptor: | C-type lectin domain family 1 member B, Snaclec rhodocytin subunit alpha, Snaclec rhodocytin subunit beta | | Authors: | Nagae, M, Morita-Matsumoto, K, Kato, M, Kato-Kaneko, M, Kato, Y, Yamaguchi, Y. | | Deposit date: | 2014-06-20 | | Release date: | 2014-10-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | A Platform of C-type Lectin-like Receptor CLEC-2 for Binding O-Glycosylated Podoplanin and Nonglycosylated Rhodocytin
Structure, 22, 2014
|
|
3WSR
 
 | | Crystal structure of CLEC-2 in complex with O-glycosylated podoplanin | | Descriptor: | C-type lectin domain family 1 member B, Peptide from Podoplanin, beta-D-galactopyranose-(1-3)-[N-acetyl-alpha-neuraminic acid-(2-6)]2-acetamido-2-deoxy-alpha-D-galactopyranose | | Authors: | Nagae, M, Morita-Matsumoto, K, Kato, M, Kato-Kaneko, M, Kato, Y, Yamaguchi, Y. | | Deposit date: | 2014-03-20 | | Release date: | 2014-10-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | A Platform of C-type Lectin-like Receptor CLEC-2 for Binding O-Glycosylated Podoplanin and Nonglycosylated Rhodocytin
Structure, 22, 2014
|
|
6FTE
 
 | | Crystal structure of an (R)-selective amine transaminase from Exophiala xenobiotica | | Descriptor: | ACETATE ION, Amine transaminase (fold IV), GLYCEROL, ... | | Authors: | Telzerow, A, Hakansson, M, Schurrmann, M, Schwab, H, Steiner, K. | | Deposit date: | 2018-02-21 | | Release date: | 2019-01-09 | | Last modified: | 2020-01-29 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Amine Transaminase from Exophiala xenobiotica - Crystal Structure and Engineering of a Fold IV Transaminase that Naturally Converts Biaryl Ketones
Acs Catalysis, 2018
|
|
1NP7
 
 | | Crystal Structure Analysis of Synechocystis sp. PCC6803 cryptochrome | | Descriptor: | DNA photolyase, FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION | | Authors: | Brudler, R, Hitomi, K, Daiyasu, H, Toh, H, Kucho, K, Ishiura, M, Kanehisa, M, Roberts, V.A, Todo, T, Tainer, J.A, Getzoff, E.D. | | Deposit date: | 2003-01-17 | | Release date: | 2003-01-28 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Identification of a new cryptochrome class: structure, function, and evolution
Mol.Cell, 11, 2003
|
|
4P6T
 
 | | Crystal Structure of tyrosinase from Bacillus megaterium with p-tyrosol in the active site | | Descriptor: | 4-(2-hydroxyethyl)phenol, COPPER (II) ION, Tyrosinase | | Authors: | Goldfeder, M, Kanteev, M, Adir, N, Fishman, A. | | Deposit date: | 2014-03-25 | | Release date: | 2014-07-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Determination of tyrosinase substrate-binding modes reveals mechanistic differences between type-3 copper proteins.
Nat Commun, 5, 2014
|
|
2EGD
 
 | | Crystal structure of human S100A13 in the Ca2+-bound state | | Descriptor: | CALCIUM ION, Protein S100-A13 | | Authors: | Imai, F.L, Nagata, K, Yonezawa, N, Nakano, M, Tanokura, M. | | Deposit date: | 2007-02-28 | | Release date: | 2008-03-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of human S100A13 in the Ca2+-bound state
Acta Crystallogr.,Sect.F, 64, 2008
|
|
4P6R
 
 | | Crystal Structure of tyrosinase from Bacillus megaterium with tyrosine in the active site | | Descriptor: | TYROSINE, Tyrosinase, ZINC ION | | Authors: | Goldfeder, M, Kanteev, M, Adir, N, Fishman, A. | | Deposit date: | 2014-03-25 | | Release date: | 2014-07-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Determination of tyrosinase substrate-binding modes reveals mechanistic differences between type-3 copper proteins.
Nat Commun, 5, 2014
|
|
4P6S
 
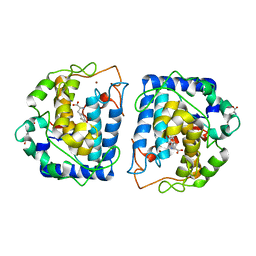 | | Crystal Structure of tyrosinase from Bacillus megaterium with L-DOPA in the active site | | Descriptor: | 3,4-DIHYDROXYPHENYLALANINE, Tyrosinase, ZINC ION | | Authors: | Goldfeder, M, Kanteev, M, Adir, N, Fishman, A. | | Deposit date: | 2014-03-25 | | Release date: | 2014-07-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Determination of tyrosinase substrate-binding modes reveals mechanistic differences between type-3 copper proteins.
Nat Commun, 5, 2014
|
|
2XG3
 
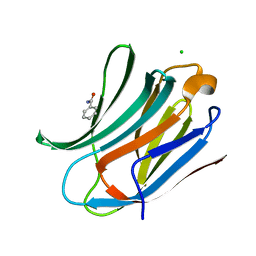 | | Human galectin-3 in complex with a benzamido-N-acetyllactoseamine inhibitor | | Descriptor: | BENZAMIDE, CHLORIDE ION, Galectin-3, ... | | Authors: | Diehl, C, Engstrom, O, Delaine, T, Hakansson, M, Genheden, S, Modig, K, Leffler, H, Ryde, U, Nilsson, U, Akke, M. | | Deposit date: | 2010-05-30 | | Release date: | 2010-10-13 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Protein flexibility and conformational entropy in ligand design targeting the carbohydrate recognition domain of galectin-3.
J. Am. Chem. Soc., 132, 2010
|
|
