2TDD
 
 | | STRUCTURES OF THYMIDYLATE SYNTHASE WITH A C-TERMINAL DELETION: ROLE OF THE C-TERMINUS IN ALIGNMENT OF D/UMP AND CH2H4FOLATE | | Descriptor: | 5-FLUORO-2'-DEOXYURIDINE-5'-MONOPHOSPHATE, 5-HYDROXYMETHYLENE-6-HYDROFOLIC ACID, THYMIDYLATE SYNTHASE | | Authors: | Perry, K.M, Carreras, C.W, Chang, L.C, Santi, D.V, Stroud, R.M. | | Deposit date: | 1993-04-05 | | Release date: | 1993-07-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structures of thymidylate synthase with a C-terminal deletion: role of the C-terminus in alignment of 2'-deoxyuridine 5'-monophosphate and 5,10-methylenetetrahydrofolate.
Biochemistry, 32, 1993
|
|
2TDM
 
 | | STRUCTURE OF THYMIDYLATE SYNTHASE | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, THYMIDYLATE SYNTHASE | | Authors: | Finer-Moore, J, Stroud, R.M. | | Deposit date: | 1997-03-28 | | Release date: | 1997-08-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Refined structures of substrate-bound and phosphate-bound thymidylate synthase from Lactobacillus casei.
J.Mol.Biol., 232, 1993
|
|
1JUJ
 
 | | Human Thymidylate Synthase Bound to dUMP and LY231514, a Pyrrolo(2,3-d)pyrimidine-based Antifolate | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, 2-{4-[2-(2-AMINO-4-OXO-4,7-DIHYDRO-3H-PYRROLO[2,3-D]PYRIMIDIN-5-YL)-ETHYL]-BENZOYLAMINO}-PENTANEDIOIC ACID, THYMIDYLATE SYNTHASE | | Authors: | Sayre, P.H, Finer-Moore, J.S, Fritz, T.A, Biermann, D, Gates, S.B, MacKellar, W.C, Patel, V.F, Stroud, R.M. | | Deposit date: | 2001-08-24 | | Release date: | 2001-09-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Multi-targeted antifolates aimed at avoiding drug resistance form covalent closed inhibitory complexes with human and Escherichia coli thymidylate synthases.
J.Mol.Biol., 313, 2001
|
|
1JU6
 
 | | Human Thymidylate Synthase Complex with dUMP and LY231514, A Pyrrolo(2,3-d)pyrimidine-based Antifolate | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, 2-{4-[2-(2-AMINO-4-OXO-4,7-DIHYDRO-3H-PYRROLO[2,3-D]PYRIMIDIN-5-YL)-ETHYL]-BENZOYLAMINO}-PENTANEDIOIC ACID, PHOSPHATE ION, ... | | Authors: | Sayre, P.H, Finer-Moore, J.S, Fritz, T.A, Biermann, D, Gates, S.B, MacKellar, W.C, Patel, V.F, Stroud, R.M. | | Deposit date: | 2001-08-23 | | Release date: | 2001-09-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Multi-targeted antifolates aimed at avoiding drug resistance form covalent closed inhibitory complexes with human and Escherichia coli thymidylate synthases.
J.Mol.Biol., 313, 2001
|
|
1LDF
 
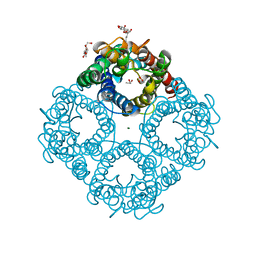 | | CRYSTAL STRUCTURE OF THE E. COLI GLYCEROL FACILITATOR (GLPF) MUTATION W48F, F200T | | Descriptor: | GLYCEROL, Glycerol uptake facilitator protein, MAGNESIUM ION, ... | | Authors: | Nollert, P, Miercke, L.J.W, O'Connell, J, Stroud, R.M. | | Deposit date: | 2002-04-08 | | Release date: | 2002-05-08 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Control of the selectivity of the aquaporin water channel family by global orientational tuning.
Science, 296, 2002
|
|
1BP0
 
 | | THYMIDYLATE SYNTHASE R23I MUTANT | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, POTASSIUM ION, PROTEIN (THYMIDYLATE SYNTHASE) | | Authors: | Morse, R, Finer-Moore, J, Stroud, R.M. | | Deposit date: | 1998-08-11 | | Release date: | 2000-01-12 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Energetic contributions of four arginines to phosphate-binding in thymidylate synthase are more than additive and depend on optimization of "effective charge balance".
Biochemistry, 39, 2000
|
|
1BO8
 
 | | THYMIDYLATE SYNTHASE R178T MUTANT | | Descriptor: | POTASSIUM ION, PROTEIN (THYMIDYLATE SYNTHASE), URIDINE-5'-MONOPHOSPHATE | | Authors: | Morse, R, Finer-Moore, J, Stroud, R.M. | | Deposit date: | 1998-08-10 | | Release date: | 1998-08-19 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Energetic contributions of four arginines to phosphate-binding in thymidylate synthase are more than additive and depend on optimization of "effective charge balance".
Biochemistry, 39, 2000
|
|
4NH0
 
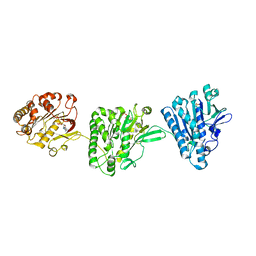 | | Cytoplasmic domain of the Thermomonospora curvata Type VII Secretion ATPase EccC | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cell divisionFtsK/SpoIIIE, MAGNESIUM ION, ... | | Authors: | Rosenberg, O.S, Cox, J.S, Stroud, R.M, Strauli, N, Dovala, D. | | Deposit date: | 2013-11-03 | | Release date: | 2015-02-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Substrates Control Multimerization and Activation of the Multi-Domain ATPase Motor of Type VII Secretion.
Cell(Cambridge,Mass.), 161, 2015
|
|
4OAA
 
 | | Crystal structure of E. coli lactose permease G46W,G262W bound to sugar | | Descriptor: | Lactose/galactose transporter, beta-D-galactopyranose-(1-1)-1-thio-beta-D-galactopyranose | | Authors: | Kumar, H, Kasho, V, Smirnova, I, Finer-Moore, J, Kaback, H.R, Stroud, R.M. | | Deposit date: | 2014-01-03 | | Release date: | 2014-01-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structure of sugar-bound LacY.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4N1A
 
 | | Thermomonospora curvata EccC (ATPases 2 and 3) in complex with a signal sequence peptide | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cell divisionFtsK/SpoIIIE, MAGNESIUM ION, ... | | Authors: | Dovala, D.L, Bendebury, A, Cox, J.S, Stroud, R.M, Rosenberg, O.S. | | Deposit date: | 2013-10-03 | | Release date: | 2015-02-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.24 Å) | | Cite: | Substrates Control Multimerization and Activation of the Multi-Domain ATPase Motor of Type VII Secretion.
Cell(Cambridge,Mass.), 161, 2015
|
|
1BP6
 
 | | THYMIDYLATE SYNTHASE R23I, R179T DOUBLE MUTANT | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, POTASSIUM ION, PROTEIN (THYMIDYLATE SYNTHASE) | | Authors: | Morse, R.J, Finer-Moore, J.S, Stroud, R.M. | | Deposit date: | 1998-08-13 | | Release date: | 1998-08-19 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Energetic contributions of four arginines to phosphate-binding in thymidylate synthase are more than additive and depend on optimization of "effective charge balance".
Biochemistry, 39, 2000
|
|
1NG1
 
 | | N AND GTPASE DOMAINS OF THE SIGNAL SEQUENCE RECOGNITION PROTEIN FFH FROM THERMUS AQUATICUS | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, CADMIUM ION, ... | | Authors: | Freymann, D.M, Stroud, R.M, Walter, P. | | Deposit date: | 1998-04-30 | | Release date: | 1999-07-30 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Functional changes in the structure of the SRP GTPase on binding GDP and Mg2+GDP.
Nat.Struct.Biol., 6, 1999
|
|
4J37
 
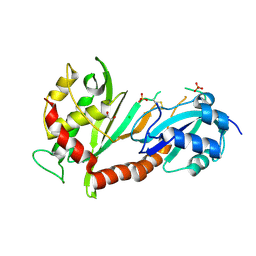 | | Crystal structure of the catalytic domain of human Pus1 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, SULFATE ION, ... | | Authors: | Czudnochowski, N, Finer-Moore, J.S, Stroud, R.M. | | Deposit date: | 2013-02-05 | | Release date: | 2013-06-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | In Human Pseudouridine Synthase 1 (hPus1), a C-Terminal Helical Insert Blocks tRNA from Binding in the Same Orientation as in the Pus1 Bacterial Homologue TruA, Consistent with Their Different Target Selectivities.
J.Mol.Biol., 425, 2013
|
|
4J6E
 
 | | Structure of LPXI D225A Mutant | | Descriptor: | (2R,3R,4R,5S,6R)-2-{[(S)-{[(S)-{[(2R,3S,4R,5R)-5-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methoxy}(hydroxy)phosphoryl]oxy}(hydroxy)phosphoryl]oxy}-5-hydroxy-6-(hydroxymethyl)-3-{[(3R)-3-hydroxytetradecanoyl]amino}tetrahydro-2H-pyran-4-yl (3R)-3-hydroxytetradecanoate, UDP-2,3-diacylglucosamine pyrophosphatase LpxI | | Authors: | Metzger IV, L.E, Lee, J.K, Finer-Moore, J.S, Raetz, C.R.H, Stroud, R.M, Center for Structures of Membrane Proteins (CSMP) | | Deposit date: | 2013-02-11 | | Release date: | 2013-05-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | LpxI structures reveal how a lipid A precursor is synthesized.
Nat.Struct.Mol.Biol., 19, 2012
|
|
4K1C
 
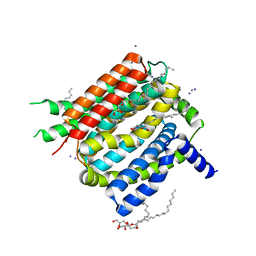 | | VCX1 Calcium/Proton Exchanger | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CALCIUM ION, IODIDE ION, ... | | Authors: | Waight, A.B, Pedersen, B.P, Stroud, R.M, Center for Structures of Membrane Proteins (CSMP) | | Deposit date: | 2013-04-04 | | Release date: | 2013-05-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for alternating access of a eukaryotic calcium/proton exchanger.
Nature, 499, 2013
|
|
4K0E
 
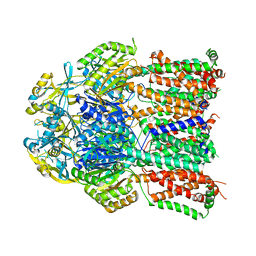 | | X-ray crystal structure of a heavy metal efflux pump, crystal form II | | Descriptor: | Heavy metal cation tricomponent efflux pump ZneA(CzcA-like), ZINC ION | | Authors: | Pak, J.E, Ngonlong Ekende, E, Vandenbussche, G, Stroud, R.M, Center for Structures of Membrane Proteins (CSMP) | | Deposit date: | 2013-04-03 | | Release date: | 2013-10-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.713 Å) | | Cite: | Structures of intermediate transport states of ZneA, a Zn(II)/proton antiporter.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4K0J
 
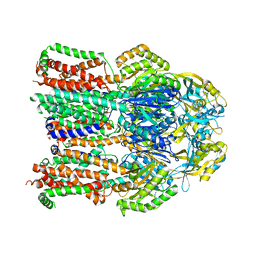 | | X-ray crystal structure of a heavy metal efflux pump, crystal form I | | Descriptor: | Heavy metal cation tricomponent efflux pump ZneA(CzcA-like), ZINC ION | | Authors: | Pak, J.E, Stroud, R.M, Ngonlong Ekende, E, Vandenbussche, G, Center for Structures of Membrane Proteins (CSMP) | | Deposit date: | 2013-04-04 | | Release date: | 2013-10-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structures of intermediate transport states of ZneA, a Zn(II)/proton antiporter.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4LGT
 
 | |
1NN6
 
 | | Human Pro-Chymase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Chymase | | Authors: | Reiling, K.K, Krucinski, J, Miercke, L.J.W, Raymond, W.W, Caughey, G.H, Stroud, R.M. | | Deposit date: | 2003-01-12 | | Release date: | 2003-03-04 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure of human pro-chymase: a model for the activating transition of granule-associated proteases.
Biochemistry, 42, 2003
|
|
4LAB
 
 | | Crystal structure of the catalytic domain of RluB | | Descriptor: | CHLORIDE ION, PLATINUM (II) ION, Ribosomal large subunit pseudouridine synthase B | | Authors: | Czudnochowski, N, Finer-Moore, J.S, Stroud, R.M. | | Deposit date: | 2013-06-19 | | Release date: | 2013-11-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5043 Å) | | Cite: | The mechanism of pseudouridine synthases from a covalent complex with RNA, and alternate specificity for U2605 versus U2604 between close homologs.
Nucleic Acids Res., 42, 2014
|
|
4JEF
 
 | |
4ITS
 
 | | Crystal structure of the catalytic domain of human Pus1 with MES in the active site | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, SULFATE ION, tRNA pseudouridine synthase A, ... | | Authors: | Czudnochowski, N, Finer-Moore, J.S, Stroud, R.M. | | Deposit date: | 2013-01-18 | | Release date: | 2013-06-05 | | Last modified: | 2013-10-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | In Human Pseudouridine Synthase 1 (hPus1), a C-Terminal Helical Insert Blocks tRNA from Binding in the Same Orientation as in the Pus1 Bacterial Homologue TruA, Consistent with Their Different Target Selectivities.
J.Mol.Biol., 425, 2013
|
|
4IQM
 
 | | Crystal structure of the catalytic domain of human Pus1 | | Descriptor: | tRNA pseudouridine synthase A, mitochondrial | | Authors: | Czudnochowski, N, Finer-Moore, J.S, Stroud, R.M. | | Deposit date: | 2013-01-11 | | Release date: | 2013-06-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | In Human Pseudouridine Synthase 1 (hPus1), a C-Terminal Helical Insert Blocks tRNA from Binding in the Same Orientation as in the Pus1 Bacterial Homologue TruA, Consistent with Their Different Target Selectivities.
J.Mol.Biol., 425, 2013
|
|
4ISK
 
 | | Crystal structure of E.coli thymidylate synthase with dUMP and the BGC 945 inhibitor | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, 2'-deoxy-5'-uridylic acid, MAGNESIUM ION, ... | | Authors: | Tochowicz, A, Finer-Moore, J, Stroud, R.M. | | Deposit date: | 2013-01-16 | | Release date: | 2013-12-25 | | Last modified: | 2014-02-12 | | Method: | X-RAY DIFFRACTION (1.752 Å) | | Cite: | Development and Binding Mode Assessment of N-[4-[2-Propyn-1-yl[(6S)-4,6,7,8-tetrahydro-2-(hydroxymethyl)-4-oxo-3H-cyclopenta[g]quinazolin-6-yl]amino]benzoyl]-l-gamma-glutamyl-d-glutamic Acid (BGC 945), a Novel Thymidylate Synthase Inhibitor That Targets Tumor Cells.
J.Med.Chem., 56, 2013
|
|
4M8B
 
 | | Fungal Protein | | Descriptor: | 1,2-ETHANEDIOL, Chain A of dsDNA containing the cis-regulatory element, Chain B of dsDNA containing the cis-regulatory element, ... | | Authors: | Rosenberg, O.S, Lohse, M.L, Stroud, R.M, Johnson, A.D. | | Deposit date: | 2013-08-13 | | Release date: | 2014-06-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Structure of a new DNA-binding domain which regulates pathogenesis in a wide variety of fungi.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
