5Y8E
 
 | |
5Y8F
 
 | |
6MOA
 
 | | C-terminal bromodomain of human BRD2 in complex with 4-(2-cyclopropyl-7-(6-methylquinolin-5-yl)-1H-benzo[d]imidazol-5-yl)-3,5-dimethylisoxazole inhibitor | | Descriptor: | 4-(2-cyclopropyl-7-(6-methylquinolin-5-yl)-1H-benzo[d]imidazol-5-yl)-3,5-dimethylisoxazole, Bromodomain-containing protein 2, GLYCEROL | | Authors: | Lansdon, E.B, Newby, Z.E.R. | | Deposit date: | 2018-10-04 | | Release date: | 2019-01-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.271 Å) | | Cite: | Structure-guided discovery of a novel, potent, and orally bioavailable 3,5-dimethylisoxazole aryl-benzimidazole BET bromodomain inhibitor.
Bioorg. Med. Chem., 27, 2019
|
|
6MO7
 
 | | N-terminal bromodomain of human BRD2 with N-((4-(3-(N-cyclopentylsulfamoyl)-4-methylphenyl)-3-methylisoxazol-5-yl)methyl)acetamide inhibitor | | Descriptor: | Bromodomain-containing protein 2, N-({4-[3-(cyclopentylsulfamoyl)-4-methylphenyl]-3-methyl-1,2-oxazol-5-yl}methyl)acetamide | | Authors: | Lansdon, E.B, Newby, Z.E.R. | | Deposit date: | 2018-10-04 | | Release date: | 2019-01-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure-guided discovery of a novel, potent, and orally bioavailable 3,5-dimethylisoxazole aryl-benzimidazole BET bromodomain inhibitor.
Bioorg. Med. Chem., 27, 2019
|
|
5ZM7
 
 | | Crystal structure of ORP1-ORD in complex with cholesterol at 3.4 A resolution | | Descriptor: | CHOLESTEROL, Oxysterol-binding protein-related protein 1 | | Authors: | Dong, J, Wang, J, Wu, J.W. | | Deposit date: | 2018-04-01 | | Release date: | 2019-02-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.401 Å) | | Cite: | Allosteric enhancement of ORP1-mediated cholesterol transport by PI(4,5)P2/PI(3,4)P2.
Nat Commun, 10, 2019
|
|
6MO9
 
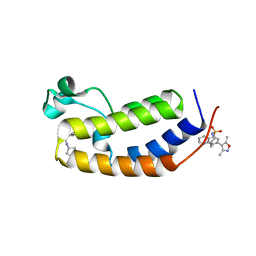 | | N-terminal bromodomain of human BRD2 in complex with N-cyclopentyl-7-(3,5-dimethylisoxazol-4-yl)quinoline-5-sulfonamide inhibitor | | Descriptor: | Bromodomain-containing protein 2, N-cyclopentyl-7-(3,5-dimethyl-1,2-oxazol-4-yl)quinoline-5-sulfonamide | | Authors: | Lansdon, E.B, Newby, Z.E.R. | | Deposit date: | 2018-10-04 | | Release date: | 2019-01-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Structure-guided discovery of a novel, potent, and orally bioavailable 3,5-dimethylisoxazole aryl-benzimidazole BET bromodomain inhibitor.
Bioorg. Med. Chem., 27, 2019
|
|
6A2W
 
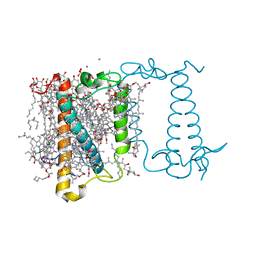 | | Crystal structure of fucoxanthin chlorophyll a/c complex from Phaeodactylum tricornutum | | Descriptor: | (3S,3'R,5R,6S,7cis)-7',8'-didehydro-5,6-dihydro-5,6-epoxy-beta,beta-carotene-3,3'-diol, (3S,3'S,5R,5'R,6S,6'R,8'R)-3,5'-dihydroxy-8-oxo-6',7'-didehydro-5,5',6,6',7,8-hexahydro-5,6-epoxy-beta,beta-caroten-3'- yl acetate, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Wang, W, Yu, L.J, Kuang, T.Y, Shen, J.R. | | Deposit date: | 2018-06-13 | | Release date: | 2019-02-06 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for blue-green light harvesting and energy dissipation in diatoms.
Science, 363, 2019
|
|
6NI2
 
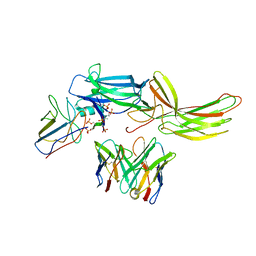 | | Stabilized beta-arrestin 1-V2T subcomplex of a GPCR-G protein-beta-arrestin mega-complex | | Descriptor: | Beta-arrestin-1, Fab30 Heavy Chain, Fab30 Light Chain, ... | | Authors: | Nguyen, A.H, Thomsen, A.R.B, Cahill, T.J, des Georges, A, Lefkowitz, R.J. | | Deposit date: | 2018-12-26 | | Release date: | 2019-11-20 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structure of an endosomal signaling GPCR-G protein-beta-arrestin megacomplex.
Nat.Struct.Mol.Biol., 26, 2019
|
|
6NI3
 
 | | B2V2R-Gs protein subcomplex of a GPCR-G protein-beta-arrestin mega-complex | | Descriptor: | 8-[(1R)-2-{[1,1-dimethyl-2-(2-methylphenyl)ethyl]amino}-1-hydroxyethyl]-5-hydroxy-2H-1,4-benzoxazin-3(4H)-one, Endolysin,Beta-2 adrenergic receptor chimera, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Nguyen, A.H, Thomsen, A.R.B, Cahill, T.J, des Georges, A, Lefkowitz, R.J. | | Deposit date: | 2018-12-26 | | Release date: | 2019-11-20 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure of an endosomal signaling GPCR-G protein-beta-arrestin megacomplex.
Nat.Struct.Mol.Biol., 26, 2019
|
|
6NNJ
 
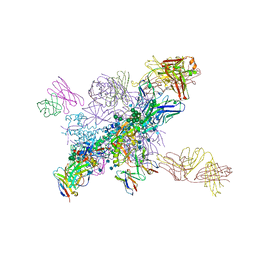 | |
6NM6
 
 | |
7YGI
 
 | | Crystal structure of p53 DBD domain in complex with azurin | | Descriptor: | Azurin, Cellular tumor antigen p53, PHOSPHATE ION, ... | | Authors: | Jiang, W.X, Zuo, J.Q, Hu, J.J, Chen, X.Q, Ma, L.X, Liu, Z, Xing, Q. | | Deposit date: | 2022-07-11 | | Release date: | 2023-02-08 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of bacterial effector protein azurin targeting tumor suppressor p53 and inhibiting its ubiquitination.
Commun Biol, 6, 2023
|
|
6MO8
 
 | | N-terminal bromodomain of human BRD2 in complex with 4,4'-(quinoline-5,7-diyl)bis(3,5-dimethylisoxazole) inhibitor | | Descriptor: | 5,7-bis(3,5-dimethyl-1,2-oxazol-4-yl)quinoline, Bromodomain-containing protein 2, SULFATE ION | | Authors: | Lansdon, E.B, Newby, Z.E.R. | | Deposit date: | 2018-10-04 | | Release date: | 2019-01-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-guided discovery of a novel, potent, and orally bioavailable 3,5-dimethylisoxazole aryl-benzimidazole BET bromodomain inhibitor.
Bioorg. Med. Chem., 27, 2019
|
|
6MWE
 
 | | CRYSTAL STRUCTURE OF TIE2 IN COMPLEX WITH DECIPERA COMPOUND DP1919 | | Descriptor: | 4-[4-({[3-tert-butyl-1-(quinolin-6-yl)-1H-pyrazol-5-yl]carbamoyl}amino)-3-fluorophenoxy]-N-methylpyridine-2-carboxamide, Angiopoietin-1 receptor, SULFATE ION | | Authors: | Chun, L, Abendroth, J, Edwards, T.E. | | Deposit date: | 2018-10-29 | | Release date: | 2018-11-21 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The Selective Tie2 Inhibitor Rebastinib Blocks Recruitment and Function of Tie2
Mol. Cancer Ther., 16, 2017
|
|
6NOX
 
 | | Solution structure of SFTI-KLK5 inhibitor | | Descriptor: | SFTI-KLK5 Peptide | | Authors: | White, A.M. | | Deposit date: | 2019-01-16 | | Release date: | 2019-04-03 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Amino Acid Scanning at P5' within the Bowman-Birk Inhibitory Loop Reveals Specificity Trends for Diverse Serine Proteases.
J. Med. Chem., 62, 2019
|
|
5ZM6
 
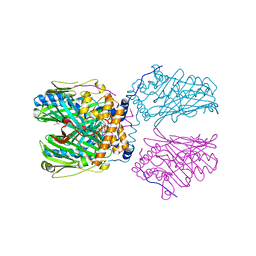 | | Crystal structure of ORP1-ORD in complex with PI(4,5)P2 | | Descriptor: | ACETATE ION, Oxysterol-binding protein-related protein 1, [(2~{S})-1-octadecanoyloxy-3-[oxidanyl-[(1~{R},2~{R},3~{S},4~{S},5~{S},6~{S})-2,3,6-tris(oxidanyl)-4,5-diphosphonooxy-cyclohexyl]oxy-phosphoryl]oxy-propan-2-yl] icosa-5,8,11,14-tetraenoate | | Authors: | Dong, J, Wang, J, Luo, Z, Wu, J.W. | | Deposit date: | 2018-04-01 | | Release date: | 2019-02-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Allosteric enhancement of ORP1-mediated cholesterol transport by PI(4,5)P2/PI(3,4)P2.
Nat Commun, 10, 2019
|
|
6NNF
 
 | |
5ZM5
 
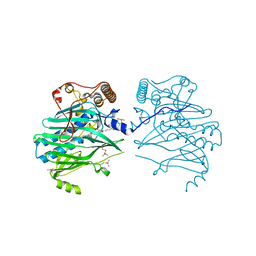 | | Crystal structure of human ORP1-ORD in complex with cholesterol at 2.6 A resolution | | Descriptor: | CHOLESTEROL, Oxysterol-binding protein-related protein 1 | | Authors: | Dong, J, Wang, J, Wu, J.W. | | Deposit date: | 2018-04-01 | | Release date: | 2019-02-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Allosteric enhancement of ORP1-mediated cholesterol transport by PI(4,5)P2/PI(3,4)P2.
Nat Commun, 10, 2019
|
|
1CKJ
 
 | |
8HPB
 
 | |
7D42
 
 | |
8HVS
 
 | |
8HQB
 
 | | NMR Structure of OsCIE1-Ubox | | Descriptor: | U-box domain-containing protein 12 | | Authors: | Zhang, Y, Yu, C.Z, Lan, W.X. | | Deposit date: | 2022-12-13 | | Release date: | 2023-12-20 | | Last modified: | 2024-06-12 | | Method: | SOLUTION NMR | | Cite: | Release of a ubiquitin brake activates OsCERK1-triggered immunity in rice.
Nature, 629, 2024
|
|
8HZM
 
 | | A new fluorescent RNA aptamer bound with N, manganese soak | | Descriptor: | (5~{Z})-5-[[4-[2-hydroxyethyl(methyl)amino]phenyl]methylidene]-3-methyl-2-[(~{E})-2-phenylethenyl]imidazol-4-one, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Huang, K.Y, Ren, A.M. | | Deposit date: | 2023-01-09 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis of a small monomeric Clivia fluorogenic RNA with a large Stokes shift.
Nat.Chem.Biol., 2024
|
|
8HZE
 
 | | A new fluorescent RNA aptamer bound with N | | Descriptor: | (5~{Z})-5-[[4-[2-hydroxyethyl(methyl)amino]phenyl]methylidene]-3-methyl-2-[(~{E})-2-phenylethenyl]imidazol-4-one, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Huang, K.Y, Ren, A.M. | | Deposit date: | 2023-01-09 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Structural basis of a small monomeric Clivia fluorogenic RNA with a large Stokes shift.
Nat.Chem.Biol., 2024
|
|
