1VR4
 
 | | Crystal Structure of MCSG TArget APC22750 from Bacillus cereus | | Descriptor: | hypothetical protein APC22750 | | Authors: | Yang, X, Brunzelle, J.S, McNamara, L.K, Minasov, G, Shuvalova, L, Collart, F.R, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-01-31 | | Release date: | 2005-02-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Crystal Structure of MCSG TArget APC22750 from Bacillus cereus
To be Published
|
|
8YLN
 
 | | The structure of DSR2-Tail tube complex | | Descriptor: | Bacillus phage SPR Tube protein, SIR2-like domain-containing protein | | Authors: | Zheng, J, Yang, X. | | Deposit date: | 2024-03-06 | | Release date: | 2024-08-14 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.53 Å) | | Cite: | Structural insights into autoinhibition and activation of defense-associated sirtuin protein.
Int.J.Biol.Macromol., 277, 2024
|
|
8Z18
 
 | |
8YLT
 
 | | The structure of DSR2 and NAD+ complex | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SIR2-like domain-containing protein | | Authors: | Zheng, J, Yang, X. | | Deposit date: | 2024-03-06 | | Release date: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (3.09 Å) | | Cite: | Structural insights into autoinhibition and activation of defense-associated sirtuin protein.
Int.J.Biol.Macromol., 277, 2024
|
|
8YKF
 
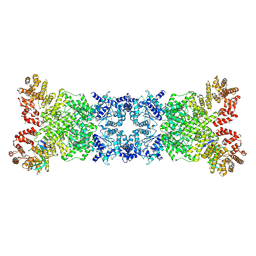 | | The DSR2-DSAD1 complex with DSAD1 on the opposite sides | | Descriptor: | DSAD1, SIR2-like domain-containing protein | | Authors: | Zheng, J, Yang, X. | | Deposit date: | 2024-03-05 | | Release date: | 2024-08-14 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | Structural insights into autoinhibition and activation of defense-associated sirtuin protein.
Int.J.Biol.Macromol., 277, 2024
|
|
8ZTR
 
 | |
3SLU
 
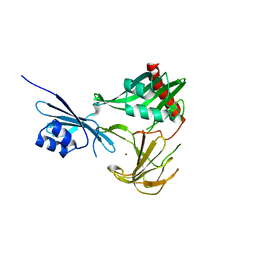 | | Crystal structure of NMB0315 | | Descriptor: | M23 peptidase domain protein, NICKEL (II) ION | | Authors: | Shen, Y, Wang, X, Yang, X, Xu, H. | | Deposit date: | 2011-06-26 | | Release date: | 2012-02-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Crystal structure of outer membrane protein NMB0315 from Neisseria meningitidis.
Plos One, 6, 2011
|
|
5IZ6
 
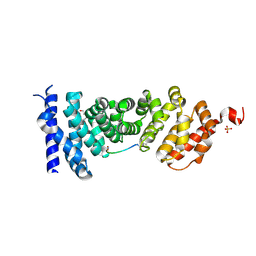 | | Protein-protein interaction | | Descriptor: | Adenomatous polyposis coli protein, DI(HYDROXYETHYL)ETHER, PHQ-ALA-GLY-GLU-ALA-LEU-TYR-GLU-NH2, ... | | Authors: | Zhao, Y, Jiang, H, Yang, X, Jiang, F, Song, K, Zhang, J. | | Deposit date: | 2016-03-25 | | Release date: | 2017-07-05 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Peptidomimetic inhibitors of APC-Asef interaction block colorectal cancer migration.
Nat. Chem. Biol., 13, 2017
|
|
9JQ1
 
 | |
3FGQ
 
 | | Crystal structure of native human neuroserpin | | Descriptor: | GLYCEROL, Neuroserpin | | Authors: | Takehara, S, Yang, X, Mikami, B, Onda, M. | | Deposit date: | 2008-12-08 | | Release date: | 2009-04-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | The 2.1-A crystal structure of native neuroserpin reveals unique structural elements that contribute to conformational instability
J.Mol.Biol., 388, 2009
|
|
6VOC
 
 | | icosahedral symmetry reconstruction of brome mosaic virus (RNA 3+4) | | Descriptor: | Capsid protein | | Authors: | Beren, C, Cui, Y.X, Chakravarty, A, Yang, X, Rao, A.L.N, Knobler, C.M, Zhou, Z.H, Gelbart, W.M. | | Deposit date: | 2020-01-30 | | Release date: | 2020-05-20 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Genome organization and interaction with capsid protein in a multipartite RNA virus.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6LAE
 
 | | Crystal structure of the DNA-binding domain of human XPA in complex with DNA | | Descriptor: | DNA (5'-D(P*GP*CP*AP*TP*CP*TP*CP*GP*CP*CP*T)-3'), DNA (5'-D(P*TP*GP*GP*CP*GP*AP*GP*AP*TP*GP*C)-3'), DNA repair protein complementing XP-A cells, ... | | Authors: | Lian, F.M, Yang, X, Jiang, Y.L, Yang, F, Li, C, Yang, W, Qian, C. | | Deposit date: | 2019-11-12 | | Release date: | 2020-02-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | New structural insights into the recognition of undamaged splayed-arm DNA with a single pair of non-complementary nucleotides by human nucleotide excision repair protein XPA.
Int.J.Biol.Macromol., 148, 2020
|
|
7F6V
 
 | | Cryo-EM structure of the human TACAN channel in a closed state | | Descriptor: | CHOLESTEROL, Ion channel TACAN | | Authors: | Chen, X.Z, Wang, Y.J, Li, Y, Yang, X, Shen, Y.Q. | | Deposit date: | 2021-06-25 | | Release date: | 2022-02-16 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.66 Å) | | Cite: | Cryo-EM structure of the human TACAN in a closed state.
Cell Rep, 38, 2022
|
|
5JMT
 
 | | Crystal structure of Zika virus NS3 helicase | | Descriptor: | NS3 helicase | | Authors: | Tian, H, Ji, X, Yang, X, Xie, W, Yang, K, Chen, C, Wu, C, Chi, H, Mu, Z, Wang, Z, Yang, H. | | Deposit date: | 2016-04-29 | | Release date: | 2016-05-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.796 Å) | | Cite: | The crystal structure of Zika virus helicase: basis for antiviral drug design
Protein Cell, 7, 2016
|
|
5X5Y
 
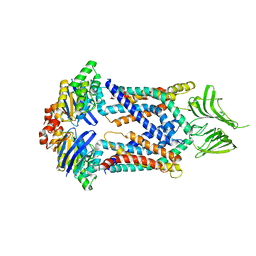 | | A membrane protein complex | | Descriptor: | Probable ATP-binding component of ABC transporter, Uncharacterized protein | | Authors: | Luo, Q, Yang, X, Huang, Y. | | Deposit date: | 2017-02-18 | | Release date: | 2017-04-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.465 Å) | | Cite: | Structural basis for lipopolysaccharide extraction by ABC transporter LptB2FG
Nat. Struct. Mol. Biol., 24, 2017
|
|
1NOQ
 
 | | e-motif structure | | Descriptor: | 5'-D(*CP*CP*GP*CP*CP*G)-3' | | Authors: | Zheng, M, Huang, X, Smith, G.K, Yang, X, Gao, X. | | Deposit date: | 2003-01-16 | | Release date: | 2003-02-11 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Genetically unstable CXG repeats are structurally dynamic and have a high propensity for folding.
An NMR and UV spectroscopic study.
J.Mol.Biol., 264, 1996
|
|
5XJX
 
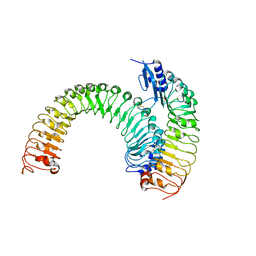 | | Pre-formed plant receptor ERL1-TMM complex | | Descriptor: | LRR receptor-like serine/threonine-protein kinase ERL1, Protein TOO MANY MOUTHS | | Authors: | Chai, J, Lin, G, Zhang, L, Han, Z, Yang, X, Liu, W, Qi, Y, Chang, J, Li, E. | | Deposit date: | 2017-05-04 | | Release date: | 2019-01-23 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.055 Å) | | Cite: | A receptor-like protein acts as a specificity switch for the regulation of stomatal development.
Genes Dev., 31, 2017
|
|
4ZM6
 
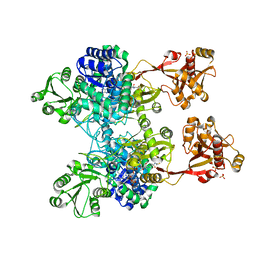 | | A unique GCN5-related glucosamine N-acetyltransferase region exist in the fungal multi-domain GH3 beta-N-acetylglucosaminidase | | Descriptor: | ACETYL COENZYME *A, N-acetyl-beta-D glucosaminidase, SULFATE ION | | Authors: | Qin, Z, Xiao, Y, Yang, X, Jiang, Z, Yang, S, Mesters, J.R. | | Deposit date: | 2015-05-02 | | Release date: | 2015-12-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A unique GCN5-related glucosamine N-acetyltransferase region exist in the fungal multi-domain glycoside hydrolase family 3 beta-N-acetylglucosaminidase
Sci Rep, 5, 2015
|
|
5XGI
 
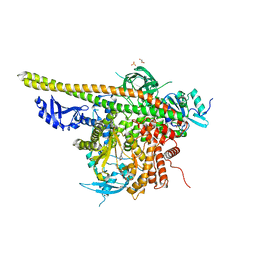 | | Crystal structure of PI3K complex with an inhibitor | | Descriptor: | 3-azanyl-5-(4-morpholin-4-ylthieno[3,2-d]pyrimidin-2-yl)phenol, GLYCEROL, HEXAETHYLENE GLYCOL, ... | | Authors: | Song, K, Yang, X, Zhao, Y, Jian, Z. | | Deposit date: | 2017-04-13 | | Release date: | 2018-04-25 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Crystal structure of PI3K complex with an inhibitor
To Be Published
|
|
5EAZ
 
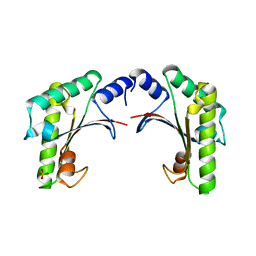 | | crystal form I of YfiB belonging to space groups P21 | | Descriptor: | SULFATE ION, YfiB | | Authors: | Xu, M, Yang, X, Yang, X.-A, Zhou, L, Liu, T.-Z, Fan, Z, Jiang, T. | | Deposit date: | 2015-10-17 | | Release date: | 2016-05-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.151 Å) | | Cite: | Structural insights into the regulatory mechanism of the Pseudomonas aeruginosa YfiBNR system
Protein Cell, 7, 2016
|
|
5EB3
 
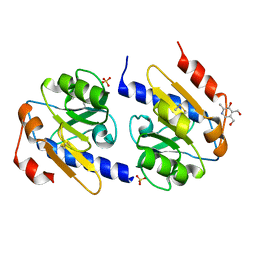 | | VB6-bound protein | | Descriptor: | 4,5-bis(hydroxymethyl)-2-methyl-pyridin-3-ol, SULFATE ION, YfiR | | Authors: | Xu, M, Yang, X, Yang, X.-A, Zhou, L, Liu, T.-Z, Fan, Z, Jiang, T. | | Deposit date: | 2015-10-17 | | Release date: | 2016-05-18 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural insights into the regulatory mechanism of the Pseudomonas aeruginosa YfiBNR system
Protein Cell, 7, 2016
|
|
5EB0
 
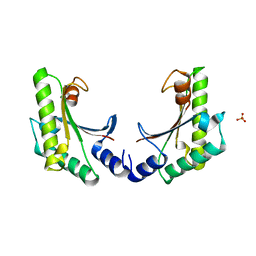 | | crystal form II of YfiB belonging to P41 | | Descriptor: | SULFATE ION, YfiB | | Authors: | Xu, M, Yang, X, Yang, X.-A, Zhou, L, Liu, T.-Z, Fan, Z, Jiang, T. | | Deposit date: | 2015-10-17 | | Release date: | 2016-05-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural insights into the regulatory mechanism of the Pseudomonas aeruginosa YfiBNR system
Protein Cell, 7, 2016
|
|
5EB2
 
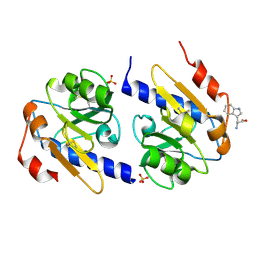 | | Trp-bound YfiR | | Descriptor: | SULFATE ION, TRYPTOPHAN, YfiR | | Authors: | Xu, M, Yang, X, Yang, X.-A, Zhou, L, Liu, T.-Z, Fan, Z, Jiang, T. | | Deposit date: | 2015-10-17 | | Release date: | 2016-05-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.709 Å) | | Cite: | Structural insights into the regulatory mechanism of the Pseudomonas aeruginosa YfiBNR system
Protein Cell, 7, 2016
|
|
5EB1
 
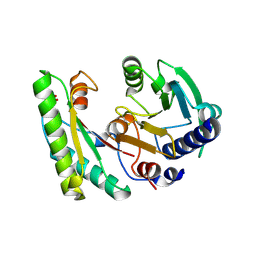 | | the YfiB-YfiR complex | | Descriptor: | SULFATE ION, YfiB, YfiR | | Authors: | Xu, M, Yang, X, Yang, X.-A, Zhou, L, Liu, T.-Z, Fan, Z, Jiang, T. | | Deposit date: | 2015-10-17 | | Release date: | 2016-05-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural insights into the regulatory mechanism of the Pseudomonas aeruginosa YfiBNR system
Protein Cell, 7, 2016
|
|
5T4D
 
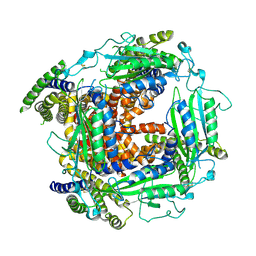 | | Cryo-EM structure of Polycystic Kidney Disease protein 2 (PKD2), residues 198-703 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, hPKD:198-703, Polycystin-2 | | Authors: | Shen, P.S, Yang, X, DeCaen, P.G, Liu, X, Bulkley, D, Clapham, D.E, Cao, E. | | Deposit date: | 2016-08-29 | | Release date: | 2016-11-02 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | The Structure of the Polycystic Kidney Disease Channel PKD2 in Lipid Nanodiscs.
Cell, 167, 2016
|
|
