1MDB
 
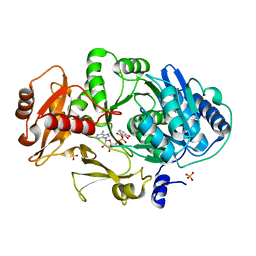 | | CRYSTAL STRUCTURE OF DhbE IN COMPLEX WITH DHB-ADENYLATE | | Descriptor: | 2,3-DIHYDROXY-BENZOIC ACID, 2,3-dihydroxybenzoate-AMP ligase, ADENOSINE MONOPHOSPHATE, ... | | Authors: | May, J.J, Kessler, N, Marahiel, M.A, Stubbs, M.T. | | Deposit date: | 2002-08-07 | | Release date: | 2002-09-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of DhbE, an archetype for aryl acid activating domains of modular nonribosomal peptide synthetases.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1Q4W
 
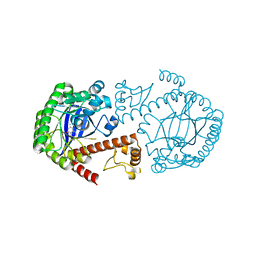 | | CRYSTAL STRUCTURE OF TGT IN COMPLEX WITH 2,6-DIAMINO-3H-QUINAZOLIN-4-ONE | | Descriptor: | 2,6-DIAMINO-3H-QUINAZOLIN-4-ONE, Queuine tRNA-ribosyltransferase, ZINC ION | | Authors: | Brenk, R, Meyer, E, Reuter, K, Stubbs, M.T, Garcia, G.A, Klebe, G. | | Deposit date: | 2003-08-04 | | Release date: | 2004-04-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystallographic Study of Inhibitors of tRNA-guanine Transglycosylase Suggests a New Structure-based Pharmacophore for Virtual Screening.
J.Mol.Biol., 338, 2004
|
|
1Q65
 
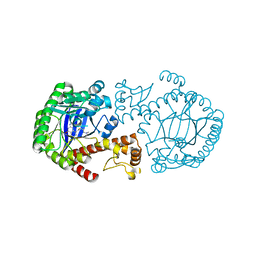 | | CRYSTAL STRUCTURE OF TGT IN COMPLEX WITH 2,6-DIAMINO-8-(2-dimethylaminoethylsulfanylmethyl)-3H-QUINAZOLIN-4-ONE crystallized at pH 5.5 | | Descriptor: | 2,6-DIAMINO-8-(2-DIMETHYLAMINOETHYLSULFANYLMETHYL)-3H-QUINAZOLIN-4-ONE, Queuine tRNA-ribosyltransferase, ZINC ION | | Authors: | Brenk, R, Meyer, E, Reuter, K, Stubbs, M.T, Garcia, G.A, Klebe, G. | | Deposit date: | 2003-08-12 | | Release date: | 2004-04-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystallographic Study of Inhibitors of tRNA-guanine Transglycosylase Suggests a New Structure-based Pharmacophore for Virtual Screening.
J.Mol.Biol., 338, 2004
|
|
1Q63
 
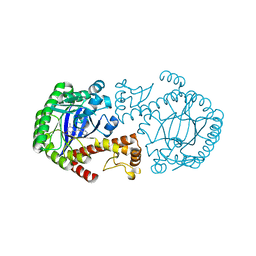 | | CRYSTAL STRUCTURE OF TGT IN COMPLEX WITH 2,6-Diamino-8-(1H-imidazol-2-ylsulfanylmethyl)-3H-quinazoline-4-one crystallized at pH 5.5 | | Descriptor: | 2,6-DIAMINO-8-(1H-IMIDAZOL-2-YLSULFANYLMETHYL)-3H-QUINAZOLINE-4-ONE, Queuine tRNA-ribosyltransferase, ZINC ION | | Authors: | Brenk, R, Meyer, E, Reuter, K, Stubbs, M.T, Garcia, G.A, Klebe, G. | | Deposit date: | 2003-08-12 | | Release date: | 2004-04-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystallographic Study of Inhibitors of tRNA-guanine Transglycosylase Suggests a New Structure-based Pharmacophore for Virtual Screening.
J.Mol.Biol., 338, 2004
|
|
1N2V
 
 | | Crystal Structure of TGT in complex with 2-Butyl-5,6-dihydro-1H-imidazo[4,5-d]pyridazine-4,7-dione | | Descriptor: | 2-BUTYL-5,6-DIHYDRO-1H-IMIDAZO[4,5-D]PYRIDAZINE-4,7-DIONE, Queuine tRNA-ribosyltransferase, ZINC ION | | Authors: | Brenk, R, Naerum, L, Graedler, U, Gerber, H.-D, Garcia, G.A, Reuter, K, Stubbs, M.T, Klebe, G. | | Deposit date: | 2002-10-24 | | Release date: | 2003-04-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Virtual screening for submicromolar leads of tRNA-guanine transglycosylase based on a new unexpected binding mode detected by crystal structure analysis
J.Med.Chem., 46, 2003
|
|
1MT0
 
 | | ATP-binding domain of hemolysin B from Escherichia coli | | Descriptor: | Hemolysin secretion ATP-binding protein, SULFATE ION | | Authors: | Schmitt, L, Benabdelhak, H, Blight, M.A, Holland, I.B, Stubbs, M.T. | | Deposit date: | 2002-09-20 | | Release date: | 2003-06-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of the nucleotide binding domain of the
ABC-transporter hemolysin B: identification of a variable region
within ABC helical domains
J.Mol.Biol., 330, 2003
|
|
1MDF
 
 | | CRYSTAL STRUCTURE OF DhbE IN ABSENCE OF SUBSTRATE | | Descriptor: | 2,3-dihydroxybenzoate-AMP ligase, SULFATE ION | | Authors: | May, J.J, Kessler, N, Marahiel, M.A, Stubbs, M.T. | | Deposit date: | 2002-08-07 | | Release date: | 2002-09-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of DhbE, an archetype for aryl acid activating domains of modular nonribosomal peptide synthetases.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
4FWU
 
 | | Crystal structure of glutaminyl cyclase from drosophila melanogaster in space group I4 | | Descriptor: | 1,2-ETHANEDIOL, CG32412, SULFATE ION, ... | | Authors: | Kolenko, P, Koch, B, Stubbs, M.T. | | Deposit date: | 2012-07-02 | | Release date: | 2012-09-05 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of glutaminyl cyclase from Drosophila melanogaster in space group I4.
Acta Crystallogr.,Sect.F, 69, 2013
|
|
4E1B
 
 | | Re-refinement of PDB entry 2EQA - SUA5 protein from Sulfolobus tokodaii with bound threonylcarbamoyladenylate | | Descriptor: | MAGNESIUM ION, YrdC/Sua5 family protein, threonylcarbamoyladenylate | | Authors: | Parthier, C, Goerlich, S, Jaenecke, F, Breithaupt, C, Braeuer, U, Fandrich, U, Clausnitzer, D, Wehmeier, U.F, Boettcher, C, Scheel, D, Stubbs, M.T. | | Deposit date: | 2012-03-06 | | Release date: | 2012-03-14 | | Last modified: | 2012-05-02 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The O-Carbamoyltransferase TobZ Catalyzes an Ancient Enzymatic Reaction.
Angew.Chem.Int.Ed.Engl., 51, 2012
|
|
4F9U
 
 | | Structure of glycosylated glutaminyl cyclase from Drosophila melanogaster | | Descriptor: | 1-(3,4-dimethoxyphenyl)-3-[3-(1H-imidazol-1-yl)propyl]thiourea, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CG32412, ... | | Authors: | Kolenko, P, Koch, B, Ruiz-Carilo, D, Stubbs, M.T. | | Deposit date: | 2012-05-21 | | Release date: | 2012-08-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structures of Glutaminyl Cyclases (QCs) from Drosophila melanogaster Reveal Active Site Conservation between Insect and Mammalian QCs.
Biochemistry, 51, 2012
|
|
4F9V
 
 | | Structure of C113A/C136A mutant variant of glycosylated glutaminyl cyclase from Drosophila melanogaster | | Descriptor: | 1-(3,4-dimethoxyphenyl)-3-[3-(1H-imidazol-1-yl)propyl]thiourea, CG32412, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kolenko, P, Koch, B, Ruiz-Carilo, D, Stubbs, M.T. | | Deposit date: | 2012-05-21 | | Release date: | 2012-08-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structures of Glutaminyl Cyclases (QCs) from Drosophila melanogaster Reveal Active Site Conservation between Insect and Mammalian QCs.
Biochemistry, 51, 2012
|
|
4FBE
 
 | | Crystal structure of the C136A/C164A variant of mitochondrial isoform of glutaminyl cyclase from Drosophila melanogaster | | Descriptor: | 1-(3,4-dimethoxyphenyl)-3-[3-(1H-imidazol-1-yl)propyl]thiourea, CG5976, isoform B, ... | | Authors: | Kolenko, P, Koch, B, Ruiz-Carilo, D, Stubbs, M.T. | | Deposit date: | 2012-05-23 | | Release date: | 2012-08-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Crystal Structures of Glutaminyl Cyclases (QCs) from Drosophila melanogaster Reveal Active Site Conservation between Insect and Mammalian QCs.
Biochemistry, 51, 2012
|
|
4HI6
 
 | |
4HIY
 
 | |
4FAI
 
 | | Crystal structure of mitochondrial isoform of glutaminyl cyclase from Drosophila melanogaster | | Descriptor: | 1-(3,4-dimethoxyphenyl)-3-[3-(1H-imidazol-1-yl)propyl]thiourea, CG5976, isoform B, ... | | Authors: | Kolenko, P, Koch, B, Ruiz-Carilo, D, Stubbs, M.T. | | Deposit date: | 2012-05-22 | | Release date: | 2012-08-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal Structures of Glutaminyl Cyclases (QCs) from Drosophila melanogaster Reveal Active Site Conservation between Insect and Mammalian QCs.
Biochemistry, 51, 2012
|
|
4HIU
 
 | |
4HIW
 
 | |
4HI5
 
 | |
4HIT
 
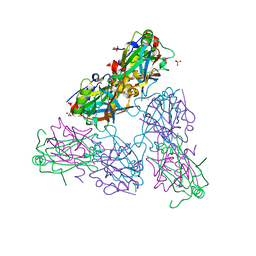 | |
3V13
 
 | | Bovine trypsin variant X(tripleGlu217Phe227) in complex with small molecule inhibitor | | Descriptor: | 3-(3-carbamimidoylphenyl)-N-(2'-sulfamoylbiphenyl-4-yl)-1,2-oxazole-4-carboxamide, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Tziridis, A, Neumann, P, Kolenko, P, Stubbs, M.T. | | Deposit date: | 2011-12-09 | | Release date: | 2012-12-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Correlating structure and ligand affinity in drug discovery: a cautionary tale involving second shell residues.
Biol.Chem., 395, 2014
|
|
3VF2
 
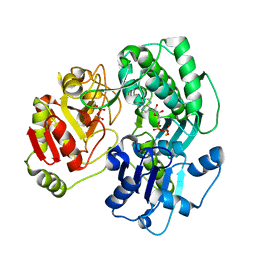 | | Crystal structure of the O-carbamoyltransferase TobZ M473I variant in complex with carbamoyl phosphate and ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, FE (II) ION, O-carbamoyltransferase TobZ, ... | | Authors: | Parthier, C, Stubbs, M.T, Goerlich, S, Jaenecke, F. | | Deposit date: | 2012-01-09 | | Release date: | 2012-01-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The O-Carbamoyltransferase TobZ Catalyzes an Ancient Enzymatic Reaction.
Angew.Chem.Int.Ed.Engl., 51, 2012
|
|
3VEZ
 
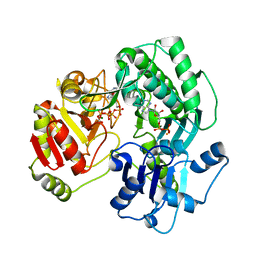 | | Crystal structure of the O-carbamoyltransferase TobZ K443A variant in complex with ATP, ADP and carbamoyl phosphate | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, FE (II) ION, ... | | Authors: | Parthier, C, Stubbs, M.T, Goerlich, S, Jaenecke, F. | | Deposit date: | 2012-01-09 | | Release date: | 2012-01-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The O-Carbamoyltransferase TobZ Catalyzes an Ancient Enzymatic Reaction.
Angew.Chem.Int.Ed.Engl., 51, 2012
|
|
3UNQ
 
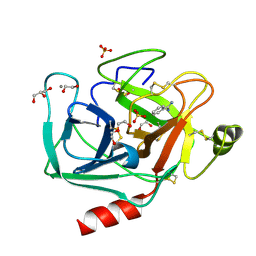 | | Bovine trypsin variant X(triplePhe227) in complex with small molecule inhibitor | | Descriptor: | 1,2-ETHANEDIOL, BENZAMIDINE, CALCIUM ION, ... | | Authors: | Tziridis, A, Neumann, P, Kolenko, P, Stubbs, M.T. | | Deposit date: | 2011-11-16 | | Release date: | 2012-11-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Correlating structure and ligand affinity in drug discovery: a cautionary tale involving second shell residues.
Biol.Chem., 395, 2014
|
|
3UNS
 
 | | Bovine trypsin variant X(triplePhe227) in complex with small molecule inhibitor | | Descriptor: | CALCIUM ION, Cationic trypsin, GLYCEROL, ... | | Authors: | Tziridis, A, Neumann, P, Kolenko, P, Stubbs, M.T. | | Deposit date: | 2011-11-16 | | Release date: | 2012-11-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Correlating structure and ligand affinity in drug discovery: a cautionary tale involving second shell residues.
Biol.Chem., 395, 2014
|
|
3UOP
 
 | | Bovine trypsin variant X(triplePhe227) in complex with small molecule inhibitor | | Descriptor: | CALCIUM ION, CHLORIDE ION, Cationic trypsin, ... | | Authors: | Tziridis, A, Neumann, P, Kolenko, P, Stubbs, M.T. | | Deposit date: | 2011-11-17 | | Release date: | 2012-11-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Correlating structure and ligand affinity in drug discovery: a cautionary tale involving second shell residues.
Biol.Chem., 395, 2014
|
|
