1ZMX
 
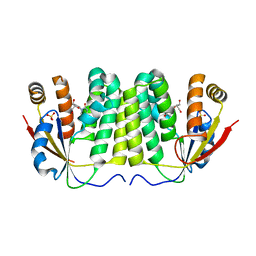 | | Crystal structure of D. melanogaster deoxyribonucleoside kinase N64D mutant in complex with thymidine | | Descriptor: | Deoxynucleoside kinase, SULFATE ION, THYMIDINE | | Authors: | Welin, M, Skovgaard, T, Knecht, W, Berenstein, D, Munch-Petersen, B, Piskur, J, Eklund, H. | | Deposit date: | 2005-05-11 | | Release date: | 2005-05-24 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural basis for the changed substrate specificity of Drosophila melanogaster deoxyribonucleoside kinase mutant N64D.
Febs J., 272, 2005
|
|
1ZM7
 
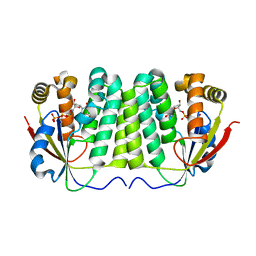 | | Crystal structure of D. melanogaster deoxyribonucleoside kinase mutant N64D in complex with dTTP | | Descriptor: | Deoxynucleoside kinase, MAGNESIUM ION, THYMIDINE-5'-TRIPHOSPHATE | | Authors: | Welin, M, Skovgaard, T, Knecht, W, Berenstein, D, Munch-Petersen, B, Piskur, J, Eklund, H. | | Deposit date: | 2005-05-10 | | Release date: | 2005-05-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for the changed substrate specificity of Drosophila melanogaster deoxyribonucleoside kinase mutant N64D.
Febs J., 272, 2005
|
|
2WRS
 
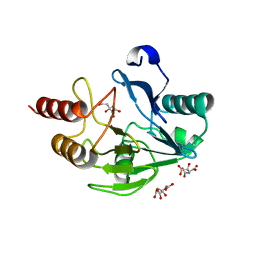 | | Crystal Structure of the Mono-Zinc Metallo-beta-lactamase VIM-4 from Pseudomonas aeruginosa | | Descriptor: | BETA-LACTAMASE VIM-4, CITRATE ANION, CITRIC ACID, ... | | Authors: | Lassaux, P, Hamel, M, Gulea, M, Delbruck, H, Traore, D.A.K, Mercuri, P.S, Horsfall, L, Dehareng, D, Gaumont, A.-C, Frere, J.-M, Ferrer, J.-L, Hoffmann, K, Galleni, M, Bebrone, C. | | Deposit date: | 2009-09-02 | | Release date: | 2010-06-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Mercaptophosphonate Compounds as Broad-Spectrum Inhibitors of the Metallo-Beta-Lactamases.
J.Med.Chem., 53, 2010
|
|
1K76
 
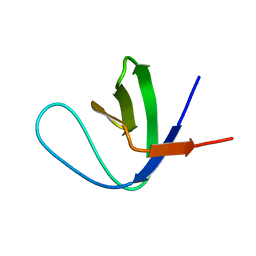 | | Solution Structure of the C-terminal Sem-5 SH3 Domain (Minimized Average Structure) | | Descriptor: | SEX MUSCLE ABNORMAL PROTEIN 5 | | Authors: | Ferreon, J, Volk, D, Luxon, B, Gorenstein, D, Hilser, V. | | Deposit date: | 2001-10-18 | | Release date: | 2003-05-20 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structure, Dynamics and Thermodynamics of the Native State Ensemble
of Sem-5 C-Terminal SH3 Domain
Biochemistry, 42, 2003
|
|
3EBA
 
 | | CAbHul6 FGLW mutant (humanized) in complex with human lysozyme | | Descriptor: | CAbHul6, Lysozyme C, SULFATE ION | | Authors: | Loris, R, Vincke, C, Saerens, D, Martinez-Rodriguez, S, Muyldermans, S, Conrath, K. | | Deposit date: | 2008-08-27 | | Release date: | 2008-12-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | General Strategy to Humanize a Camelid Single-domain Antibody and Identification of a Universal Humanized Nanobody Scaffold
J.Biol.Chem., 284, 2009
|
|
3EAK
 
 | | NbBCII10 humanized (FGLA mutant) | | Descriptor: | NbBCII10-FGLA, SULFATE ION | | Authors: | Vincke, C, Loris, R, Saerens, D, Martinez-Rodriguez, S, Muyldermans, S, Conrath, K. | | Deposit date: | 2008-08-26 | | Release date: | 2008-12-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | General strategy to humanize a camelid single-domain antibody and identification of a universal humanized nanobody scaffold.
J.Biol.Chem., 2008
|
|
3EUL
 
 | | Structure of the signal receiver domain of the putative response regulator NarL from Mycobacterium tuberculosis | | Descriptor: | CHLORIDE ION, POSSIBLE NITRATE/NITRITE RESPONSE TRANSCRIPTIONAL REGULATORY PROTEIN NARL (DNA-binding response regulator, LuxR family) | | Authors: | Schneider, G, Schnell, R, Agren, D. | | Deposit date: | 2008-10-10 | | Release date: | 2008-11-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | 1.9 A structure of the signal receiver domain of the putative response regulator NarL from Mycobacterium tuberculosis.
Acta Crystallogr.,Sect.F, 64, 2008
|
|
1KFZ
 
 | | Solution Structure of C-terminal Sem-5 SH3 Domain (Ensemble of 16 Structures) | | Descriptor: | SEX MUSCLE ABNORMAL PROTEIN 5 | | Authors: | Ferreon, J.C, Volk, D.E, Luxon, B.A, Gorenstein, D, Hilser, V.J. | | Deposit date: | 2001-11-24 | | Release date: | 2003-05-20 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structure, Dynamics and Thermodynamics of the Native State Ensemble
of Sem-5 C-Terminal SH3 Domain
Biochemistry, 42, 2003
|
|
1J3G
 
 | | Solution structure of Citrobacter Freundii AmpD | | Descriptor: | AmpD protein, ZINC ION | | Authors: | Liepinsh, E, Genereux, C, Dehareng, D, Joris, B, Otting, G. | | Deposit date: | 2003-01-31 | | Release date: | 2003-02-18 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of Citrobacter freundii AmpD, Comparison with Bacteriophage T7 Lysozyme
and Homology with PGRP Domains
J.Mol.Biol., 327, 2003
|
|
1E3Y
 
 | |
1E41
 
 | |
1CXR
 
 | |
2JZI
 
 | |
7V8N
 
 | |
1C54
 
 | | SOLUTION STRUCTURE OF RIBONUCLEASE SA | | Descriptor: | RIBONUCLEASE SA | | Authors: | Laurents, D.V, Canadillas-Perez, J.M, Santoro, J, Schell, D, Pace, C.N, Rico, M, Bruix, M. | | Deposit date: | 1999-10-22 | | Release date: | 2001-11-28 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure and dynamics of ribonuclease Sa.
Proteins, 44, 2001
|
|
2M0K
 
 | |
2M0J
 
 | |
2L4V
 
 | |
1BCG
 
 | | SCORPION TOXIN BJXTR-IT | | Descriptor: | TOXIN BJXTR-IT | | Authors: | Oren, D, Froy, O, Amit, E, Kleinberger-Doron, N, Gurevitz, M, Shaanan, B. | | Deposit date: | 1998-04-29 | | Release date: | 1998-11-18 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | An excitatory scorpion toxin with a distinctive feature: an additional alpha helix at the C terminus and its implications for interaction with insect sodium channels.
Structure, 6, 1998
|
|
4G9C
 
 | | Human B-Raf Kinase Domain bound to a Type II Pyrazolopyridine Inhibitor | | Descriptor: | 3-{[3-(2-cyanopropan-2-yl)benzoyl]amino}-2,6-difluoro-N-(3-methoxy-2H-pyrazolo[3,4-b]pyridin-5-yl)benzamide, Serine/threonine-protein kinase B-raf | | Authors: | Voegtli, W.C, Sturgis, H.L. | | Deposit date: | 2012-07-23 | | Release date: | 2012-11-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Pyrazolopyridine inhibitors of B-Raf(V600E). Part 4: Rational design and kinase selectivity profile of cell potent type II inhibitors.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
7DE7
 
 | | Crystal structure of PDZD7 HHD domain | | Descriptor: | (2R)-2-{[(2R)-2-{[(2S)-2-{[(2R)-2-hydroxypropyl]oxy}propyl]oxy}propyl]oxy}propan-1-ol, PDZ domain-containing protein 7 | | Authors: | Wang, H, Lin, L, Lu, Q. | | Deposit date: | 2020-11-02 | | Release date: | 2021-08-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Structure and Membrane Targeting of the PDZD7 Harmonin Homology Domain (HHD) Associated With Hearing Loss.
Front Cell Dev Biol, 9, 2021
|
|
5K4J
 
 | | Crystal Structure of CDK2 in complex with compound 22 | | Descriptor: | 1-[(1~{S})-1-(4-chloranyl-3-fluoranyl-phenyl)-2-oxidanyl-ethyl]-4-[2-[(2-methylpyrazol-3-yl)amino]pyrimidin-4-yl]pyridin-2-one, Cyclin-dependent kinase 2 | | Authors: | Yin, J, Wang, W. | | Deposit date: | 2016-05-20 | | Release date: | 2016-07-06 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Discovery of (S)-1-(1-(4-Chloro-3-fluorophenyl)-2-hydroxyethyl)-4-(2-((1-methyl-1H-pyrazol-5-yl)amino)pyrimidin-4-yl)pyridin-2(1H)-one (GDC-0994), an Extracellular Signal-Regulated Kinase 1/2 (ERK1/2) Inhibitor in Early Clinical Development.
J.Med.Chem., 59, 2016
|
|
5K4I
 
 | | Crystal Structure of ERK2 in complex with compound 22 | | Descriptor: | 1,2-ETHANEDIOL, 1-[(1~{S})-1-(4-chloranyl-3-fluoranyl-phenyl)-2-oxidanyl-ethyl]-4-[2-[(2-methylpyrazol-3-yl)amino]pyrimidin-4-yl]pyridin-2-one, Mitogen-activated protein kinase 1 | | Authors: | Yin, J, Wang, W. | | Deposit date: | 2016-05-20 | | Release date: | 2016-07-06 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Discovery of (S)-1-(1-(4-Chloro-3-fluorophenyl)-2-hydroxyethyl)-4-(2-((1-methyl-1H-pyrazol-5-yl)amino)pyrimidin-4-yl)pyridin-2(1H)-one (GDC-0994), an Extracellular Signal-Regulated Kinase 1/2 (ERK1/2) Inhibitor in Early Clinical Development.
J.Med.Chem., 59, 2016
|
|
6NKO
 
 | | Crystal structure of ForH | | Descriptor: | ForH | | Authors: | Zheng, J, Irani, S, Zhang, Y. | | Deposit date: | 2019-01-07 | | Release date: | 2019-04-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.403 Å) | | Cite: | Identification of the Formycin A Biosynthetic Gene Cluster from Streptomyces kaniharaensis Illustrates the Interplay between Biological Pyrazolopyrimidine Formation and de Novo Purine Biosynthesis.
J. Am. Chem. Soc., 141, 2019
|
|
1ES4
 
 | | C98N mutant of streptomyces K15 DD-transpeptidase | | Descriptor: | DD-TRANSPEPTIDASE | | Authors: | Fonze, E, Charlier, P. | | Deposit date: | 2000-04-07 | | Release date: | 2000-05-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Catalytic mechanism of the Streptomyces K15 DD-transpeptidase/penicillin-binding protein probed by site-directed mutagenesis and structural analysis.
Biochemistry, 42, 2003
|
|
