8HYM
 
 | | Crystal structure of AtHPPD-Y18030 complex | | Descriptor: | 3-cyclopentyl-1,5-dimethyl-6-(2-oxidanyl-6-oxidanylidene-cyclohexen-1-yl)carbonyl-quinazoline-2,4-dione, 4-hydroxyphenylpyruvate dioxygenase, COBALT (II) ION | | Authors: | Dong, J, Lin, H.-Y, Yang, G.-F. | | Deposit date: | 2023-01-07 | | Release date: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.794 Å) | | Cite: | Crystal structure of AtHPPD-Y18030 complex
To Be Published
|
|
8I15
 
 | | Crystal structure of AtHPPD-YH20335 complex | | Descriptor: | 1,4-dimethyl-5-(2-oxidanyl-6-oxidanylidene-cyclohexen-1-yl)carbonyl-3-(phenylmethyl)benzimidazol-2-one, 4-hydroxyphenylpyruvate dioxygenase, COBALT (II) ION | | Authors: | Dong, J, Lin, H.-Y, Yang, G.-F. | | Deposit date: | 2023-01-12 | | Release date: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Crystal structure of AtHPPD-YH20335 complex
To Be Published
|
|
8I2U
 
 | | Crystal structure of AtHPPD-YH20282 complex | | Descriptor: | 2-[2-chloranyl-4-methylsulfonyl-3-(2-trimethylsilylethoxy)phenyl]carbonyl-3-oxidanyl-cyclohex-2-en-1-one, 4-hydroxyphenylpyruvate dioxygenase, COBALT (II) ION | | Authors: | Dong, J, Lin, H.-Y, Yang, G.-F. | | Deposit date: | 2023-01-15 | | Release date: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.989 Å) | | Cite: | Crystal structure of AtHPPD-YH20282 complex
To Be Published
|
|
8I2S
 
 | | Crystal structure of AtHPPD-Y18979 complex | | Descriptor: | 1,5-dimethyl-3-(naphthalen-2-ylmethyl)-6-(2-oxidanyl-6-oxidanylidene-cyclohexen-1-yl)carbonyl-quinazoline-2,4-dione, 4-hydroxyphenylpyruvate dioxygenase, COBALT (II) ION | | Authors: | Dong, J, Lin, H.-Y, Yang, G.-F. | | Deposit date: | 2023-01-15 | | Release date: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.592 Å) | | Cite: | Crystal structure of AtHPPD-Y18979 complex
To Be Published
|
|
8HX2
 
 | | Crystal structure of AtHPPD-Y18405 complex | | Descriptor: | 3-[2-(3-chlorophenyl)ethyl]-1,5-dimethyl-6-(2-oxidanyl-6-oxidanylidene-cyclohexa-1,3-dien-1-yl)carbonyl-quinazoline-2,4-dione, 4-hydroxyphenylpyruvate dioxygenase, COBALT (II) ION | | Authors: | Dong, J, Lin, H.-Y, Yang, G.-F. | | Deposit date: | 2023-01-03 | | Release date: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.996 Å) | | Cite: | Crystal structure of AtHPPD-Y18405 complex
To Be Published
|
|
8HZ9
 
 | | Crystal structure of AtHPPD-Y181136 complex | | Descriptor: | 4-hydroxyphenylpyruvate dioxygenase, 5-methyl-6-[(2-methyl-3-oxidanylidene-1H-pyrazol-4-yl)carbonyl]-3-propan-2-yl-1,2,3-benzotriazin-4-one, COBALT (II) ION | | Authors: | Dong, J, Lin, H.-Y, Yang, G.-F. | | Deposit date: | 2023-01-08 | | Release date: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.011 Å) | | Cite: | Crystal structure of AtHPPD-Y181136 complex
To Be Published
|
|
8HZ6
 
 | | Crystal structure of AtHPPD-QRY2089 complex | | Descriptor: | 1,5-dimethyl-6-(2-oxidanyl-6-oxidanylidene-cyclohexen-1-yl)carbonyl-3-prop-2-ynyl-quinazoline-2,4-dione, 4-hydroxyphenylpyruvate dioxygenase, COBALT (II) ION | | Authors: | Dong, J, Lin, H.-Y, Yang, G.-F. | | Deposit date: | 2023-01-08 | | Release date: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.605 Å) | | Cite: | Crystal structure of AtHPPD-QRY2089 complex
To Be Published
|
|
4XBL
 
 | |
4XBN
 
 | |
4XBQ
 
 | |
4V98
 
 | | The 8S snRNP Assembly Intermediate | | Descriptor: | CG10419, Icln, LD23602p, ... | | Authors: | Grimm, C, Pelz, J.P, Schindelin, H, Diederichs, K, Kuper, J, Kisker, C. | | Deposit date: | 2012-05-15 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural Basis of Assembly Chaperone- Mediated snRNP Formation.
Mol.Cell, 49, 2013
|
|
5H9P
 
 | | Crystal Structure of Human Galectin-3 CRD in Complex with TD139 | | Descriptor: | 3-deoxy-3-[4-(3-fluorophenyl)-1H-1,2,3-triazol-1-yl]-beta-D-galactopyranosyl 3-deoxy-3-[4-(3-fluorophenyl)-1H-1,2,3-triazol-1-yl]-1-thio-beta-D-galactopyranoside, Galectin-3 | | Authors: | Hsieh, T.J, Lin, H.Y, Lin, C.H. | | Deposit date: | 2015-12-29 | | Release date: | 2016-06-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Dual thio-digalactoside-binding modes of human galectins as the structural basis for the design of potent and selective inhibitors
Sci Rep, 6, 2016
|
|
5H9R
 
 | | Crystal Structure of Human Galectin-3 CRD in Complex with TAZTDG | | Descriptor: | (2~{S},3~{R},4~{S},5~{R},6~{R})-2-[(2~{S},3~{R},4~{S},5~{R},6~{R})-4-[4-(3-fluorophenyl)-1,2,3-triazol-1-yl]-6-(hydroxymethyl)-3,5-bis(oxidanyl)oxan-2-yl]sulfanyl-6-(hydroxymethyl)oxane-3,4,5-triol, Galectin-3 | | Authors: | Hsieh, T.J, Lin, H.Y, Lin, C.H. | | Deposit date: | 2015-12-29 | | Release date: | 2016-06-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Dual thio-digalactoside-binding modes of human galectins as the structural basis for the design of potent and selective inhibitors
Sci Rep, 6, 2016
|
|
5H9S
 
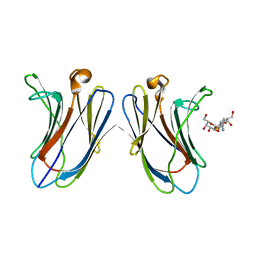 | | Crystal Structure of Human Galectin-7 in Complex with TAZTDG | | Descriptor: | (2~{S},3~{R},4~{S},5~{R},6~{R})-2-[(2~{S},3~{R},4~{S},5~{R},6~{R})-4-[4-(3-fluorophenyl)-1,2,3-triazol-1-yl]-6-(hydroxymethyl)-3,5-bis(oxidanyl)oxan-2-yl]sulfanyl-6-(hydroxymethyl)oxane-3,4,5-triol, Galectin-7 | | Authors: | Hsieh, T.J, Lin, H.Y, Lin, C.H. | | Deposit date: | 2015-12-29 | | Release date: | 2016-06-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.821 Å) | | Cite: | Dual thio-digalactoside-binding modes of human galectins as the structural basis for the design of potent and selective inhibitors
Sci Rep, 6, 2016
|
|
5ANY
 
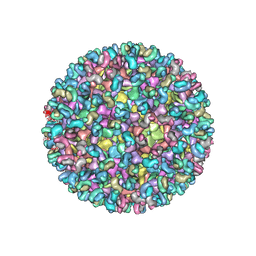 | | Electron cryo-microscopy of chikungunya virus in complex with neutralizing antibody Fab CHK265 | | Descriptor: | E1, E2, FAB, ... | | Authors: | Fox, J.M, Long, F, Edeling, M.A, Lin, H, Duijl-Richter, M, Fong, R.H, Kahle, K.M, Smit, J.M, Jin, J, Simmons, G, Doranz, B.J, Crowe, J.E, Fremont, D.H, Rossmann, M.G, Diamond, M.S. | | Deposit date: | 2015-09-08 | | Release date: | 2015-11-25 | | Last modified: | 2018-10-03 | | Method: | ELECTRON MICROSCOPY (16.9 Å) | | Cite: | Broadly Neutralizing Alphavirus Antibodies Bind an Epitope on E2 and Inhibit Entry and Egress.
Cell(Cambridge,Mass.), 163, 2015
|
|
5H9Q
 
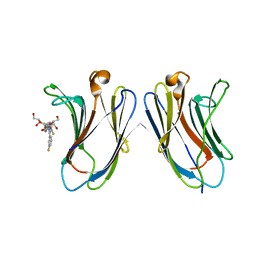 | | Crystal Structure of Human Galectin-7 in Complex with TD139 | | Descriptor: | 3-deoxy-3-[4-(3-fluorophenyl)-1H-1,2,3-triazol-1-yl]-beta-D-galactopyranosyl 3-deoxy-3-[4-(3-fluorophenyl)-1H-1,2,3-triazol-1-yl]-1-thio-beta-D-galactopyranoside, Galectin-7 | | Authors: | Hsieh, T.J, Lin, H.Y, Lin, C.H. | | Deposit date: | 2015-12-29 | | Release date: | 2016-06-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.931 Å) | | Cite: | Dual thio-digalactoside-binding modes of human galectins as the structural basis for the design of potent and selective inhibitors
Sci Rep, 6, 2016
|
|
2IO4
 
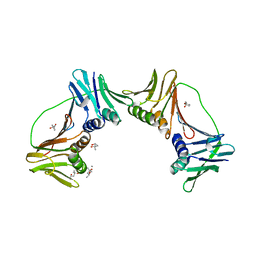 | | Crystal structure of PCNA12 dimer from Sulfolobus solfataricus. | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, DNA polymerase sliding clamp B, ... | | Authors: | Hlinkova, V, Ling, H. | | Deposit date: | 2006-10-09 | | Release date: | 2008-04-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structures of monomeric, dimeric and trimeric PCNA: PCNA-ring assembly and opening.
Acta Crystallogr.,Sect.D, 64, 2008
|
|
3E3I
 
 | |
6PAV
 
 | |
8HOW
 
 | | Crystal structure of AtHPPD-Y191052 complex | | Descriptor: | 1,5-dimethyl-6-(2-oxidanyl-6-oxidanylidene-cyclohexen-1-yl)carbonyl-3-(2-phenylethyl)quinazoline-2,4-dione, 4-hydroxyphenylpyruvate dioxygenase, COBALT (II) ION | | Authors: | Yang, G.-F, Lin, H.-Y, Dong, J. | | Deposit date: | 2022-12-11 | | Release date: | 2023-01-25 | | Last modified: | 2023-02-01 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Discovery of Subnanomolar Inhibitors of 4-Hydroxyphenylpyruvate Dioxygenase via Structure-Based Rational Design.
J.Agric.Food Chem., 71, 2023
|
|
2IA6
 
 | | Bypass of Major Benzopyrene-dG Adduct by Y-Family DNA Polymerase with Unique Structural Gap | | Descriptor: | 1,2,3-TRIHYDROXY-1,2,3,4-TETRAHYDROBENZO[A]PYRENE, 1,2-ETHANEDIOL, 5'-D(*GP*GP*GP*GP*GP*AP*AP*GP*GP*AP*TP*TP*A)-3', ... | | Authors: | Bauer, J, Ling, H, Sayer, J.M, Xing, G, Yagi, H, Jerina, D.M. | | Deposit date: | 2006-09-07 | | Release date: | 2007-09-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A structural gap in Dpo4 supports mutagenic bypass of a major benzo[a]pyrene dG adduct in DNA through template misalignment.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
5GKB
 
 | | Crystal Structure of Fatty Acid-Binding Protein in Brain Tissue of Drosophila melanogaster without citrate inside | | Descriptor: | Fatty acid bindin protein, isoform B | | Authors: | Cheng, Y.-Y, Huang, Y.-F, Lin, H.-H, Chang, W.W, Lyu, P.-C. | | Deposit date: | 2016-07-04 | | Release date: | 2017-07-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | The ligand-mediated affinity of brain-type fatty acid-binding protein for membranes determines the directionality of lipophilic cargo transport.
Biochim Biophys Acta Mol Cell Biol Lipids, 1864, 2019
|
|
5GGE
 
 | | Fatty Acid-Binding Protein in Brain Tissue of Drosophila melanogaster | | Descriptor: | CITRIC ACID, Fatty acid bindin protein, isoform B | | Authors: | Cheng, Y.-Y, Huang, Y.-F, Lin, H.-H, Chang, W.W, Lyu, P.-C. | | Deposit date: | 2016-06-15 | | Release date: | 2017-06-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.861 Å) | | Cite: | The ligand-mediated affinity of brain-type fatty acid-binding protein for membranes determines the directionality of lipophilic cargo transport.
Biochim Biophys Acta Mol Cell Biol Lipids, 1864, 2019
|
|
3E3G
 
 | |
6DO3
 
 | | KLHDC2 ubiquitin ligase in complex with SelK C-end degron | | Descriptor: | Kelch domain-containing protein 2, SelK C-end Degron | | Authors: | Rusnac, D.V, Lin, H.C, Yen, H.C.S, Zheng, N. | | Deposit date: | 2018-06-08 | | Release date: | 2018-12-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.165 Å) | | Cite: | Recognition of the Diglycine C-End Degron by CRL2KLHDC2Ubiquitin Ligase.
Mol. Cell, 72, 2018
|
|
