4ZLI
 
 | | Cellobionic acid phosphorylase - 3-O-beta-D-glucopyranosyl-alpha-D-glucopyranuronic acid complex | | Descriptor: | CHLORIDE ION, GLYCEROL, Putative b-glycan phosphorylase, ... | | Authors: | Nam, Y.W, Arakawa, T, Fushinobu, S. | | Deposit date: | 2015-05-01 | | Release date: | 2015-06-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure and Substrate Recognition of Cellobionic Acid Phosphorylase, Which Plays a Key Role in Oxidative Cellulose Degradation by Microbes.
J.Biol.Chem., 290, 2015
|
|
4ZLE
 
 | | Cellobionic acid phosphorylase - ligand free structure | | Descriptor: | CHLORIDE ION, GLYCEROL, Putative b-glycan phosphorylase, ... | | Authors: | Nam, Y.W, Arakawa, T, Fushinobu, S. | | Deposit date: | 2015-05-01 | | Release date: | 2015-06-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure and Substrate Recognition of Cellobionic Acid Phosphorylase, Which Plays a Key Role in Oxidative Cellulose Degradation by Microbes.
J.Biol.Chem., 290, 2015
|
|
5OGR
 
 | | Structure of cathepsin B1 from Schistosoma mansoni in complex with WRR286 inhibitor | | Descriptor: | 3-[[N-[4-METHYL-PIPERAZINYL]CARBONYL]-PHENYLALANINYL-AMINO]-5-PHENYL-PENTANE-1-SULFONIC ACID BENZYLOXY-AMIDE, ACETATE ION, Cathepsin B-like peptidase (C01 family) | | Authors: | Jilkova, A, Rezacova, P, Brynda, J, Mares, M. | | Deposit date: | 2017-07-13 | | Release date: | 2018-11-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Druggable Hot Spots in the Schistosomiasis Cathepsin B1 Target Identified by Functional and Binding Mode Analysis of Potent Vinyl Sulfone Inhibitors.
Acs Infect Dis., 2020
|
|
5OGQ
 
 | | Structure of cathepsin B1 from Schistosoma mansoni in complex with WRR391 inhibitor | | Descriptor: | ACETATE ION, Cathepsin B-like peptidase (C01 family), ethyl 1-[[(2~{S})-3-(4-hydroxyphenyl)-1-oxidanylidene-1-[[(3~{S})-1-phenyl-5-pyridin-2-ylsulfonyl-pentan-3-yl]amino]propan-2-yl]carbamoyl]piperidine-4-carboxylate | | Authors: | Jilkova, A, Rezacova, P, Brynda, J, Mares, M. | | Deposit date: | 2017-07-13 | | Release date: | 2018-11-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Druggable Hot Spots in the Schistosomiasis Cathepsin B1 Target Identified by Functional and Binding Mode Analysis of Potent Vinyl Sulfone Inhibitors.
Acs Infect Dis., 2020
|
|
7R7N
 
 | |
7R6X
 
 | | SARS-CoV-2 spike receptor-binding domain (RBD) in complex with S2E12 Fab, S309 Fab, and S304 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Monoclonal antibody S2E12 Fab heavy chain, ... | | Authors: | Snell, G, Czudnochowski, N, Croll, T.I, Nix, J.C, Corti, D, Cameroni, E, Pinto, D, Beltramello, M. | | Deposit date: | 2021-06-23 | | Release date: | 2021-07-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | SARS-CoV-2 RBD antibodies that maximize breadth and resistance to escape.
Nature, 597, 2021
|
|
7R6W
 
 | | SARS-CoV-2 spike receptor-binding domain (RBD) in complex with S2X35 Fab and S309 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, GLYCEROL, ... | | Authors: | Snell, G, Czudnochowski, N, Hernandez, P, Nix, J.C, Croll, T.I, Corti, D, Cameroni, E, Pinto, D, Beltramello, M. | | Deposit date: | 2021-06-23 | | Release date: | 2021-07-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | SARS-CoV-2 RBD antibodies that maximize breadth and resistance to escape.
Nature, 597, 2021
|
|
6QBG
 
 | | Crystal structure of human cathepsin D in complex with macrocyclic inhibitor 14 | | Descriptor: | (3~{S},7~{S},8~{S})-8-(naphthalen-2-ylmethyl)-7-oxidanyl-3-propan-2-yl-1,4,9-triazacyclohenicosane-2,5,10-trione, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Brynda, J, Houstecka, R, Majer, P, Mares, M. | | Deposit date: | 2018-12-21 | | Release date: | 2020-01-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Biomimetic Macrocyclic Inhibitors of Human Cathepsin D: Structure-Activity Relationship and Binding Mode Analysis.
J.Med.Chem., 63, 2020
|
|
6QBH
 
 | | Crystal structure of human cathepsin D in complex with macrocyclic inhibitor 33 | | Descriptor: | (4~{S},5~{S},9~{S})-5-oxidanyl-4-(phenylmethyl)-9-propan-2-yl-1-oxa-3,8,11-triazacyclodocosane-2,7,10-trione, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Brynda, J, Houstecka, R, Majer, P, Mares, M. | | Deposit date: | 2018-12-21 | | Release date: | 2020-01-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Biomimetic Macrocyclic Inhibitors of Human Cathepsin D: Structure-Activity Relationship and Binding Mode Analysis.
J.Med.Chem., 63, 2020
|
|
6QCB
 
 | | Crystal structure of human cathepsin D in complex with macrocyclic inhibitor 9 | | Descriptor: | (3~{S},7~{S},8~{S})-7-oxidanyl-8-(phenylmethyl)-3-propan-2-yl-1,4,9-triazacyclohenicosane-2,5,10-trione, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Cathepsin D, ... | | Authors: | Brynda, J, Houstecka, R, Majer, P, Mares, M. | | Deposit date: | 2018-12-27 | | Release date: | 2020-01-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Biomimetic Macrocyclic Inhibitors of Human Cathepsin D: Structure-Activity Relationship and Binding Mode Analysis.
J.Med.Chem., 63, 2020
|
|
5ZFI
 
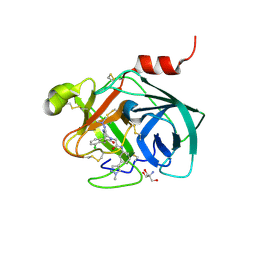 | | Mouse kallikrein 7 in complex with 6-benzyl-1,4-diazepan-7-one derivative | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-[(3Z,6R)-6-[(2,6-dichlorophenyl)methyl]-3-(dimethylhydrazinylidene)-7-oxo-1,4-diazepan-1-yl]-N-[3-(1-methyl-1H-pyrazol-4-yl)phenyl]acetamide, Kallikrein-7 | | Authors: | Sugawara, H. | | Deposit date: | 2018-03-06 | | Release date: | 2018-06-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-based drug design to overcome species differences in kallikrein 7 inhibition of 1,3,6-trisubstituted 1,4-diazepan-7-ones.
Bioorg. Med. Chem., 26, 2018
|
|
5ZFH
 
 | | Mouse Kallikrein 7 | | Descriptor: | Kallikrein-7 | | Authors: | Sugawara, H. | | Deposit date: | 2018-03-06 | | Release date: | 2018-06-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structure-based drug design to overcome species differences in kallikrein 7 inhibition of 1,3,6-trisubstituted 1,4-diazepan-7-ones.
Bioorg. Med. Chem., 26, 2018
|
|
5H4S
 
 | |
5X17
 
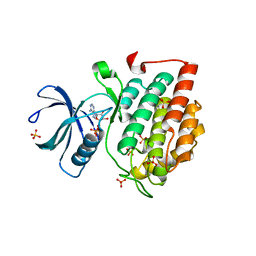 | | Crystal structure of murine CK1d in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Casein kinase I isoform delta, SULFATE ION | | Authors: | Kikuchi, M, Shinohara, Y, Ueda, H.R, Umehara, T. | | Deposit date: | 2017-01-25 | | Release date: | 2017-10-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Temperature-Sensitive Substrate and Product Binding Underlie Temperature-Compensated Phosphorylation in the Clock
Mol. Cell, 67, 2017
|
|
5X18
 
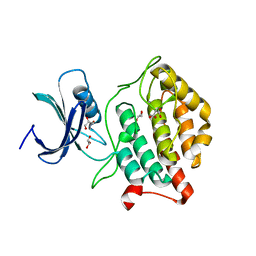 | | Crystal structure of Casein kinase I homolog 1 | | Descriptor: | Casein kinase I homolog 1, GLYCEROL, MALONIC ACID | | Authors: | Kikuchi, M, Shinohara, Y, Ueda, H.R, Umehara, T. | | Deposit date: | 2017-01-25 | | Release date: | 2017-10-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Temperature-Sensitive Substrate and Product Binding Underlie Temperature-Compensated Phosphorylation in the Clock
Mol. Cell, 67, 2017
|
|
5TF9
 
 | |
5GZK
 
 | | Endo-beta-1,2-glucanase from Chitinophaga pinensis - sophorotriose and glucose complex | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, PENTAETHYLENE GLYCOL, ... | | Authors: | Abe, K, Nakajima, M, Arakawa, T, Fushinobu, S, Taguchi, H. | | Deposit date: | 2016-09-28 | | Release date: | 2017-03-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Biochemical and structural analyses of a bacterial endo-beta-1,2-glucanase reveal a new glycoside hydrolase family
J. Biol. Chem., 292, 2017
|
|
5GZH
 
 | | Endo-beta-1,2-glucanase from Chitinophaga pinensis - ligand free form | | Descriptor: | Endo-beta-1,2-glucanase, IODIDE ION, alpha-D-glucopyranose-(1-1)-alpha-D-glucopyranose | | Authors: | Abe, K, Nakajima, M, Arakawa, T, Fushinobu, S, Taguchi, H. | | Deposit date: | 2016-09-28 | | Release date: | 2017-03-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Biochemical and structural analyses of a bacterial endo-beta-1,2-glucanase reveal a new glycoside hydrolase family
J. Biol. Chem., 292, 2017
|
|
8YBE
 
 | | Cryo-EM structure of Maltose Binding Protein | | Descriptor: | Maltose/maltodextrin-binding periplasmic protein, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Yoo, Y, Park, K, Kim, H. | | Deposit date: | 2024-02-13 | | Release date: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Atomic resolution structure of MBP using Cryo-EM
To Be Published
|
|
4ZLG
 
 | | Cellobionic acid phosphorylase - gluconic acid complex | | Descriptor: | CHLORIDE ION, D-gluconic acid, D-glucono-1,5-lactone, ... | | Authors: | Nam, Y.W, Arakawa, T, Fushinobu, S. | | Deposit date: | 2015-05-01 | | Release date: | 2015-06-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal Structure and Substrate Recognition of Cellobionic Acid Phosphorylase, Which Plays a Key Role in Oxidative Cellulose Degradation by Microbes.
J.Biol.Chem., 290, 2015
|
|
8CD9
 
 | | Cathepsin B1 from Schistosoma mansoni in complex with gallinamide analog 6 | | Descriptor: | 1,2-ETHANEDIOL, Cathepsin B-like peptidase (C01 family), SODIUM ION, ... | | Authors: | Rubesova, P, Brynda, J, Fanfrlik, J, Gerwick, W.H, Mares, M. | | Deposit date: | 2023-01-30 | | Release date: | 2024-02-21 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Nature-Inspired Gallinamides Are Potent Antischistosomal Agents: Inhibition of the Cathepsin B1 Protease Target and Binding Mode Analysis.
Acs Infect Dis., 10, 2024
|
|
8C25
 
 | | purine nucleoside phosphorylase in complex with JS-375 | | Descriptor: | CHLORIDE ION, GLYCEROL, Purine nucleoside phosphorylase, ... | | Authors: | Djukic, S, Pachl, P, Rezacova, P. | | Deposit date: | 2022-12-21 | | Release date: | 2023-05-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Design, Synthesis, Biological Evaluation, and Crystallographic Study of Novel Purine Nucleoside Phosphorylase Inhibitors.
J.Med.Chem., 66, 2023
|
|
7K45
 
 | |
8CC2
 
 | | Cathepsin B1 from Schistosoma mansoni in complex with gallinamide A | | Descriptor: | Cathepsin B-like peptidase (C01 family), SODIUM ION, gallinamide A, ... | | Authors: | Rubesova, P, Brynda, J, Mares, M, Gerwick, W.H. | | Deposit date: | 2023-01-26 | | Release date: | 2024-02-07 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Nature-Inspired Gallinamides Are Potent Antischistosomal Agents: Inhibition of the Cathepsin B1 Protease Target and Binding Mode Analysis.
Acs Infect Dis., 10, 2024
|
|
8CCU
 
 | | Cathepsin B1 from Schistosoma mansoni in complex with gallinamide analog 1 | | Descriptor: | Cathepsin B-like peptidase (C01 family), SODIUM ION, [(2~{S})-1-[[(2~{S})-1-[[(2~{S})-5-[(2~{S})-3-methoxy-2-(2-methylpropyl)-5-oxidanylidene-2~{H}-pyrrol-1-yl]-5-oxidanylidene-pentan-2-yl]amino]-4-methyl-1-oxidanylidene-pentan-2-yl]amino]-4-methyl-1-oxidanylidene-pentan-2-yl] (2~{S},3~{S})-2-(dimethylamino)-3-methyl-pentanoate | | Authors: | Rubesova, P, Brynda, J, Gerwick, W.H, Mares, M. | | Deposit date: | 2023-01-27 | | Release date: | 2024-02-07 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Nature-Inspired Gallinamides Are Potent Antischistosomal Agents: Inhibition of the Cathepsin B1 Protease Target and Binding Mode Analysis.
Acs Infect Dis., 10, 2024
|
|
