1ZNV
 
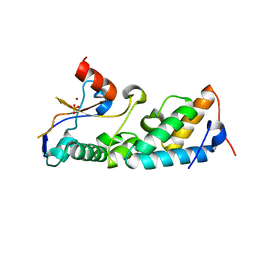 | | How a His-metal finger endonuclease ColE7 binds and cleaves DNA with a transition metal ion cofactor | | Descriptor: | Colicin E7, Colicin E7 immunity protein, NICKEL (II) ION, ... | | Authors: | Doudeva, L.G, Huang, H, Hsia, K.C, Shi, Z, Li, C.L, Shen, Y, Yuan, H.S. | | Deposit date: | 2005-05-12 | | Release date: | 2006-03-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structural analysis and metal-dependent stability and activity studies of the ColE7 endonuclease domain in complex with DNA/Zn2+ or inhibitor/Ni2+
Protein Sci., 15, 2006
|
|
8GW4
 
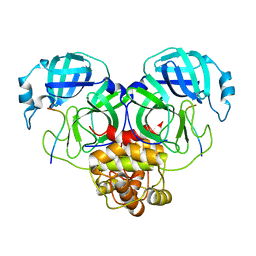 | | SARS-CoV-2 Mpro 1-302/C145A in complex with peptide 8-1 | | Descriptor: | Replicase polyprotein 1ab, peptide 8-1 | | Authors: | Liu, M, Huang, H. | | Deposit date: | 2022-09-16 | | Release date: | 2023-08-23 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The S1'-S3' Pocket of the SARS-CoV-2 Main Protease Is Critical for Substrate Selectivity and Can Be Targeted with Covalent Inhibitors.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
8GWS
 
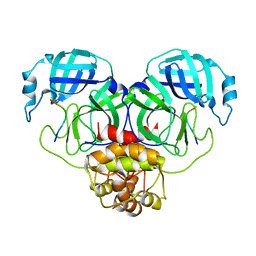 | | SARS-CoV-2 Mpro 1-302 c145a in complex with peptide 4 | | Descriptor: | Replicase polyprotein 1ab, VAL-LYS-LEU-GLN-ALA-ILE-PHE-ARG | | Authors: | Liu, M, Fu, Z, Huang, H. | | Deposit date: | 2022-09-17 | | Release date: | 2023-08-23 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The S1'-S3' Pocket of the SARS-CoV-2 Main Protease Is Critical for Substrate Selectivity and Can Be Targeted with Covalent Inhibitors.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
1ZNS
 
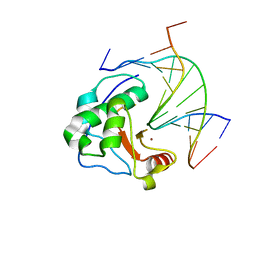 | | Crystal structure of N-ColE7/12-bp DNA/Zn complex | | Descriptor: | 5'-D(*CP*GP*GP*GP*AP*TP*AP*TP*CP*CP*CP*G)-3', Colicin E7, ZINC ION | | Authors: | Doudeva, L.G, Huang, H, Hsia, K.C, Shi, Z, Li, C.L, Shen, Y, Yuan, H.S. | | Deposit date: | 2005-05-12 | | Release date: | 2006-03-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structural analysis and metal-dependent stability and activity studies of the ColE7 endonuclease domain in complex with DNA/Zn2+ or inhibitor/Ni2+
Protein Sci., 15, 2006
|
|
2AX5
 
 | | Solution Structure of Urm1 from Saccharomyces Cerevisiae | | Descriptor: | Hypothetical 11.0 kDa protein in FAA3-MAS3 intergenic region | | Authors: | Xu, J, Huang, H, Zhang, J, Wu, J, Shi, Y. | | Deposit date: | 2005-09-03 | | Release date: | 2006-06-27 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of Urm1 and its implications for the origin of protein modifiers.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
6U8K
 
 | | Crystal structure of hepatitis C virus IRES junction IIIabc in complex with Fab HCV3 | | Descriptor: | Heavy chain of Fab HCV3, JIIIabc RNA (68-MER), Light chain of Fab HCV3 | | Authors: | Koirala, D, Lewicka, A, Koldobskaya, Y, Huang, H, Piccirilli, J.A. | | Deposit date: | 2019-09-05 | | Release date: | 2019-12-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Synthetic Antibody Binding to a Preorganized RNA Domain of Hepatitis C Virus Internal Ribosome Entry Site Inhibits Translation.
Acs Chem.Biol., 15, 2020
|
|
6M12
 
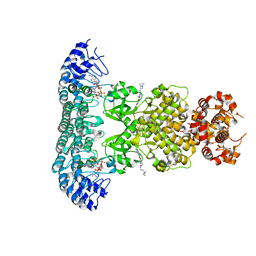 | | Crystal Structure of Rnase L in complex with SU11652 | | Descriptor: | 5-[(E)-(5-CHLORO-2-OXO-1,2-DIHYDRO-3H-INDOL-3-YLIDENE)METHYL]-N-[2-(DIETHYLAMINO)ETHYL]-2,4-DIMETHYL-1H-PYRROLE-3-CARBOXAMIDE, PHOSPHATE ION, Ribonuclease L, ... | | Authors: | Tang, J, Huang, H. | | Deposit date: | 2020-02-24 | | Release date: | 2020-09-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Sunitinib inhibits RNase L by destabilizing its active dimer conformation.
Biochem.J., 477, 2020
|
|
5IJC
 
 | | The crystal structure of mouse TLR4/MD-2/neoseptin-3 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Lymphocyte antigen 96, ... | | Authors: | Wang, Y, Su, L, Morin, M.D, Jones, B.T, Whitby, L.R, Surakattula, M, Huang, H, Shi, H, Choi, J.H, Wang, K, Moresco, E.M, Berger, M, Zhan, X, Zhang, H, Boger, D.L, Beutler, B. | | Deposit date: | 2016-03-01 | | Release date: | 2016-04-27 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | TLR4/MD-2 activation by a synthetic agonist with no similarity to LPS.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5IJB
 
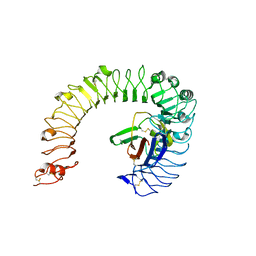 | | The ligand-free structure of the mouse TLR4/MD-2 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Lymphocyte antigen 96, ... | | Authors: | Wang, Y, Su, L, Morin, M.D, Jones, B.T, Whitby, L.R, Surakattula, M, Huang, H, Shi, H, Choi, J.H, Wang, K, Moresco, E.M, Berger, M, Zhan, X, Zhang, H, Boger, D.L, Beutler, B. | | Deposit date: | 2016-03-01 | | Release date: | 2016-04-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | TLR4/MD-2 activation by a synthetic agonist with no similarity to LPS.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5IJD
 
 | | The crystal structure of mouse TLR4/MD-2/lipid A complex | | Descriptor: | (R)-((2R,3S,4R,5R,6R)-3-HYDROXY-2-(HYDROXYMETHYL)-5-((R)-3-HYDROXYTETRADECANAMIDO)-6-(PHOSPHONOOXY)TETRAHYDRO-2H-PYRAN-4-YL) 3-HYDROXYTETRADECANOATE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wang, Y, Su, L, Morin, M.D, Jones, B.T, Whitby, L.R, Surakattula, M, Huang, H, Shi, H, Choi, J.H, Wang, K, Moresco, E.M, Berger, M, Zhan, X, Zhang, H, Boger, D.L, Beutler, B. | | Deposit date: | 2016-03-01 | | Release date: | 2016-04-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | TLR4/MD-2 activation by a synthetic agonist with no similarity to LPS.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
6U8D
 
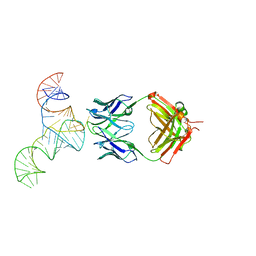 | | Crystal structure of hepatitis C virus IRES junction IIIabc in complex with Fab HCV2 | | Descriptor: | Heavy chain of Fab HCV2, JIIIabc RNA (68-MER), Light chain of Fab HCV2 | | Authors: | Koirala, D, Lewicka, A, Koldobskaya, Y, Huang, H, Piccirilli, J.A. | | Deposit date: | 2019-09-04 | | Release date: | 2019-12-04 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.807 Å) | | Cite: | Synthetic Antibody Binding to a Preorganized RNA Domain of Hepatitis C Virus Internal Ribosome Entry Site Inhibits Translation.
Acs Chem.Biol., 15, 2020
|
|
6M13
 
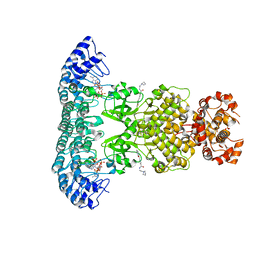 | | Crystal structure of Rnase L in complex with Toceranib | | Descriptor: | 5-[(Z)-(5-fluoranyl-2-oxidanylidene-1H-indol-3-ylidene)methyl]-2,4-dimethyl-N-(2-pyrrolidin-1-ylethyl)-1H-pyrrole-3-carboxamide, PHOSPHATE ION, Ribonuclease L, ... | | Authors: | Tang, J, Huang, H. | | Deposit date: | 2020-02-24 | | Release date: | 2020-09-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Sunitinib inhibits RNase L by destabilizing its active dimer conformation.
Biochem.J., 477, 2020
|
|
3R45
 
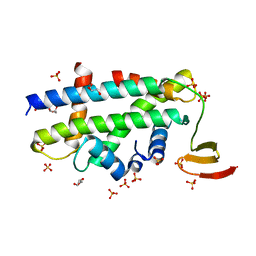 | | Structure of a CENP-A-Histone H4 Heterodimer in complex with chaperone HJURP | | Descriptor: | GLYCEROL, Histone H3-like centromeric protein A, Histone H4, ... | | Authors: | Hu, H, Liu, Y, Wang, M, Fang, J, Huang, H, Yang, N, Li, Y, Wang, J, Yao, X, Shi, Y, Li, G, Xu, R.M. | | Deposit date: | 2011-03-17 | | Release date: | 2011-04-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of a CENP-A-histone H4 heterodimer in complex with chaperone HJURP
Genes Dev., 25, 2011
|
|
9CU4
 
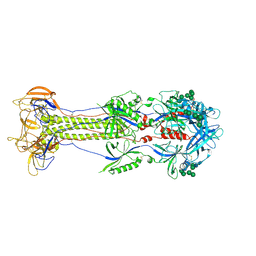 | | KSHV glycoprotein B ectodomain, postfusion form | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ORF8, ... | | Authors: | Ito, F, Zhen, J, Xie, G, Huang, H, Silva, J.C, Wu, T, Zhou, Z.H. | | Deposit date: | 2024-07-25 | | Release date: | 2025-01-08 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure of the Kaposi's sarcoma-associated herpesvirus gB in post-fusion conformation.
J.Virol., 99, 2025
|
|
3GYP
 
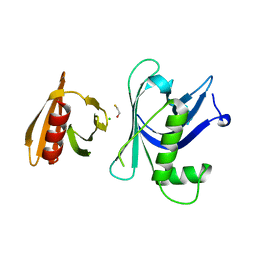 | | Rtt106p | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, Histone chaperone RTT106 | | Authors: | Liu, Y, Huang, H, Shi, Y, Teng, M. | | Deposit date: | 2009-04-04 | | Release date: | 2009-12-08 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.406 Å) | | Cite: | Structural analysis of Rtt106p reveals a DNA-binding role required for heterochromatin silencing
J.Biol.Chem., 285, 2010
|
|
3GYO
 
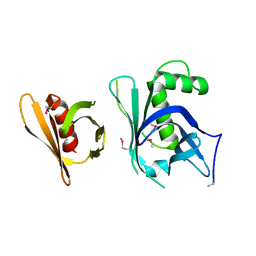 | | Se-Met Rtt106p | | Descriptor: | Histone chaperone RTT106 | | Authors: | Liu, Y, Huang, H, Shi, Y, Teng, M. | | Deposit date: | 2009-04-04 | | Release date: | 2009-12-08 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural analysis of Rtt106p reveals a DNA-binding role required for heterochromatin silencing
J.Biol.Chem., 285, 2010
|
|
2OS6
 
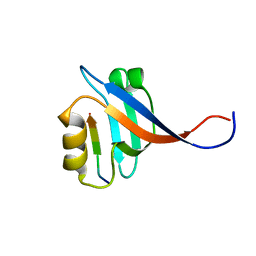 | |
4A4S
 
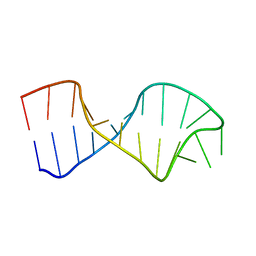 | | UNAC Tetraloops: To What Extent Can They Mimic GNRA Tetraloops | | Descriptor: | 5'-R(*GP*GP*AP*CP*CP*CP*GP*GP*CP*UP*CP*AP*CP*GP *CP*UP*GP*GP*GP*UP*CP*C)-3' | | Authors: | Zhao, Q, Huang, H, Nagaswamy, U, Xia, Y, Gao, X, Fox, G. | | Deposit date: | 2011-10-20 | | Release date: | 2012-05-30 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Unac Tetraloops: To What Extent Can They Mimic Gnra Tetraloops
Biopolymers, 97, 2012
|
|
4AX8
 
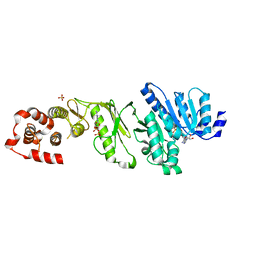 | | Medium resolution structure of the bifunctional kinase- methyltransferase WbdD | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, S-ADENOSYLMETHIONINE, ... | | Authors: | Hagelueken, G, Huang, H, Naismith, J.H. | | Deposit date: | 2012-06-11 | | Release date: | 2012-10-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystallization, Dehydration and Experimental Phasing of Wbdd, a Bifunctional Kinase and Methyltransferase from Escherichia Coli O9A.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4A4R
 
 | | UNAC Tetraloops: To What Extent Can They Mimic GNRA Tetraloops | | Descriptor: | 5'-R(*GMP*GP*AP*CP*CP*CP*GP*GP*CP*UP*AP*AP*CP*GP *CP*UP*GP*GP*GP*UP*CP*C)-3' | | Authors: | Zhao, Q, Huang, H, Nagaswamy, U, Xia, Y, Gao, X, Fox, G. | | Deposit date: | 2011-10-20 | | Release date: | 2012-05-30 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Unac Tetraloops: To What Extent Can They Mimic Gnra Tetraloops
Biopolymers, 97, 2012
|
|
4AZW
 
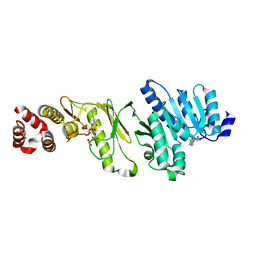 | | Crystal structure of monomeric WbdD. | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, S-ADENOSYLMETHIONINE, ... | | Authors: | Hagelueken, G, Huang, H, Naismith, J.H. | | Deposit date: | 2012-06-26 | | Release date: | 2012-09-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Structure of Wbdd; a Bifunctional Kinase and Methyltransferase that Regulates the Chain Length of the O Antigen in Escherichia Coli O9A.
Mol.Microbiol., 86, 2012
|
|
4AZT
 
 | | Co-crystal structure of WbdD and kinase inhibitor LY294002. | | Descriptor: | 2-MORPHOLIN-4-YL-7-PHENYL-4H-CHROMEN-4-ONE, CHLORIDE ION, METHYLTRANSFERASE WBDD, ... | | Authors: | Hagelueken, G, Huang, H, Naismith, J.H. | | Deposit date: | 2012-06-26 | | Release date: | 2012-09-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Structure of Wbdd; a Bifunctional Kinase and Methyltransferase that Regulates the Chain Length of the O Antigen in Escherichia Coli O9A.
Mol.Microbiol., 86, 2012
|
|
4A4T
 
 | | UNAC Tetraloops: To What Extent Can They Mimic GNRA Tetraloops | | Descriptor: | 5'-R(*GP*GP*AP*CP*CP*CP*GP*GP*CP*UP*UP*AP*CP*GP *CP*UP*GP*GP*GP*UP*CP*C)-3' | | Authors: | Zhao, Q, Huang, H, Nagaswamy, U, Xia, Y, Gao, X, Fox, G. | | Deposit date: | 2011-10-20 | | Release date: | 2012-05-30 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Unac Tetraloops: To What Extent Can They Mimic Gnra Tetraloops
Biopolymers, 97, 2012
|
|
4AZS
 
 | | High resolution (2.2 A) crystal structure of WbdD. | | Descriptor: | ADENOSINE MONOPHOSPHATE, CHLORIDE ION, METHYLTRANSFERASE WBDD, ... | | Authors: | Hagelueken, G, Huang, H, Naismith, J.H. | | Deposit date: | 2012-06-26 | | Release date: | 2012-09-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure of Wbdd; a Bifunctional Kinase and Methyltransferase that Regulates the Chain Length of the O Antigen in Escherichia Coli O9A.
Mol.Microbiol., 86, 2012
|
|
2OMJ
 
 | | solution structure of LARG PDZ domain | | Descriptor: | Rho guanine nucleotide exchange factor 12 | | Authors: | Liu, J, Huang, H, Hu, Q. | | Deposit date: | 2007-01-22 | | Release date: | 2008-01-22 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | solution structure and dynamics of the LARG PDZ domain
To be Published
|
|
