3D8T
 
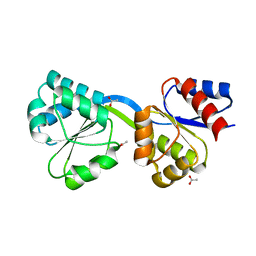 | | Thermus thermophilus Uroporphyrinogen III Synthase | | Descriptor: | ACETATE ION, Uroporphyrinogen-III synthase | | Authors: | Schubert, H.L. | | Deposit date: | 2008-05-23 | | Release date: | 2008-08-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure and mechanistic implications of a uroporphyrinogen III synthase-product complex.
Biochemistry, 47, 2008
|
|
3D8S
 
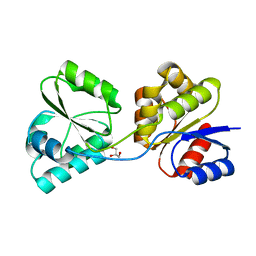 | | Thermus thermophilus Uroporphyrinogen III Synthase | | Descriptor: | GLYCEROL, Uroporphyrinogen-III synthase | | Authors: | Schubert, H.L. | | Deposit date: | 2008-05-23 | | Release date: | 2008-08-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and mechanistic implications of a uroporphyrinogen III synthase-product complex.
Biochemistry, 47, 2008
|
|
9FT8
 
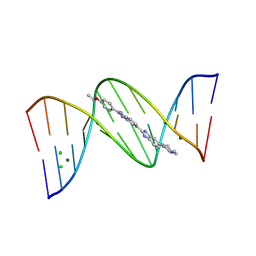 | | Crystal structure of d(CGTGAATTCACG) with HT1 | | Descriptor: | 2'-(4-ETHOXYPHENYL)-5-(4-METHYL-1-PIPERAZINYL)-2,5'-BI-BENZIMIDAZOLE, CHLORIDE ION, DNA (5'-D(*CP*GP*TP*GP*AP*AP*TP*TP*CP*AP*CP*G)-3'), ... | | Authors: | Sbirkova-Dimitrova, H.I, Shivachev, B.L. | | Deposit date: | 2024-06-24 | | Release date: | 2025-01-22 | | Method: | X-RAY DIFFRACTION (1.902 Å) | | Cite: | Structural Characterization of B-DNA d(CGTGAATTCACG)2 in Complex with the Specific Minor Groove Binding Fluorescent Marker Hoechst 33342
Crystals, 15, 2025
|
|
6XPC
 
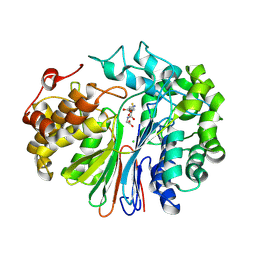 | | Structure of human GGT1 in complex with full GSH molecule | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, GLUTATHIONE, ... | | Authors: | Terzyan, S.S, Hanigan, M. | | Deposit date: | 2020-07-08 | | Release date: | 2020-11-25 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Crystal structures of glutathione- and inhibitor-bound human GGT1: critical interactions within the cysteinylglycine binding site.
J.Biol.Chem., 296, 2020
|
|
6XPB
 
 | |
5V4Q
 
 | | Crystal Structure of human GGT1 in complex with DON | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 5,5-dihydroxy-L-norleucine, CHLORIDE ION, ... | | Authors: | Terzyan, S, Hanigan, M. | | Deposit date: | 2017-03-10 | | Release date: | 2017-04-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of 6-diazo-5-oxo-norleucine-bound human gamma-glutamyl transpeptidase 1, a novel mechanism of inactivation.
Protein Sci., 26, 2017
|
|
5UKI
 
 | |
4DFA
 
 | | Crystal structure of Staphylococcal nuclease variant Delta+PHS I92A/L36A at cryogenic temperature | | Descriptor: | CALCIUM ION, THYMIDINE-3',5'-DIPHOSPHATE, Thermonuclease | | Authors: | Caro, J.A, Clark, I.A, Schlessman, J.L, Heroux, A, Garcia-Moreno E, B. | | Deposit date: | 2012-01-23 | | Release date: | 2012-02-01 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.404 Å) | | Cite: | Pressure effects in proteins
To be Published
|
|
4DGZ
 
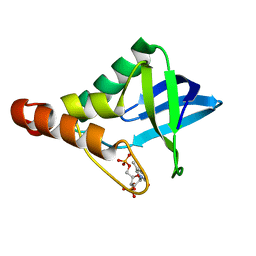 | | Crystal structure of Staphylococcal nuclease variant Delta+PHS I92A/L125A at cryogenic temperature | | Descriptor: | CALCIUM ION, THYMIDINE-3',5'-DIPHOSPHATE, Thermonuclease | | Authors: | Caro, J.A, Nam, S.P, Schlessman, J.L, Heroux, A, Garcia-Moreno E, B. | | Deposit date: | 2012-01-27 | | Release date: | 2012-02-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Pressure effects in proteins
To be Published
|
|
7ALI
 
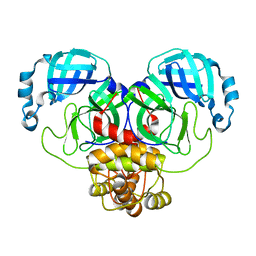 | | Crystal structure of the main protease (3CLpro/Mpro) of SARS-CoV-2 at 1.65A resolution (spacegroup P2(1)). | | Descriptor: | 3C-like proteinase | | Authors: | Costanzi, E, Demitri, N, Giabbai, B, Heroux, A, Storici, P. | | Deposit date: | 2020-10-06 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural and Biochemical Analysis of the Dual Inhibition of MG-132 against SARS-CoV-2 Main Protease (Mpro/3CLpro) and Human Cathepsin-L.
Int J Mol Sci, 22, 2021
|
|
7ALH
 
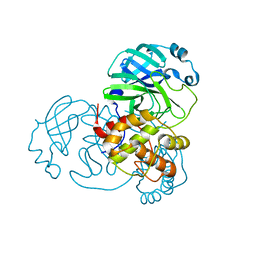 | | Crystal structure of the main protease (3CLpro/Mpro) of SARS-CoV-2 at 1.65A resolution (spacegroup C2). | | Descriptor: | 3C-like proteinase | | Authors: | Costanzi, E, Demitri, N, Giabbai, B, Heroux, A, Storici, P. | | Deposit date: | 2020-10-06 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural and Biochemical Analysis of the Dual Inhibition of MG-132 against SARS-CoV-2 Main Protease (Mpro/3CLpro) and Human Cathepsin-L.
Int J Mol Sci, 22, 2021
|
|
1Z3Y
 
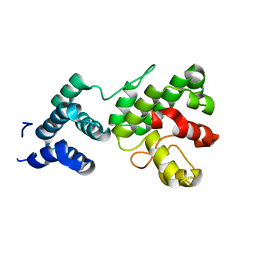 | | Structure of Gun4-1 from Thermosynechococcus elongatus | | Descriptor: | putative cytidylyltransferase | | Authors: | Davison, P.A, Schubert, H.L, Reid, J.D, Iorg, C.D, Robinson, H, Hill, C.P, Hunter, C.N. | | Deposit date: | 2005-03-14 | | Release date: | 2005-06-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and Biochemical Characterization of Gun4 Suggests a Mechanism for Its Role in Chlorophyll Biosynthesis(,).
Biochemistry, 44, 2005
|
|
1Z3X
 
 | | Structure of Gun4 from Thermosynechococcus elongatus | | Descriptor: | putative cytidylyltransferase | | Authors: | Davison, P.A, Schubert, H.L, Reid, J.D, Iorg, C.D, Robinson, H, Hill, C.P, Hunter, C.N. | | Deposit date: | 2005-03-14 | | Release date: | 2005-06-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural and Biochemical Characterization of Gun4 Suggests a Mechanism for Its Role in Chlorophyll Biosynthesis(,).
Biochemistry, 44, 2005
|
|
3F6P
 
 | | Crystal Structure of unphosphorelated receiver domain of YycF | | Descriptor: | Transcriptional regulatory protein yycF | | Authors: | Zhao, H, Tang, L. | | Deposit date: | 2008-11-06 | | Release date: | 2010-03-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Preliminary crystallographic studies of the regulatory domain of response regulator YycF from an essential two-component signal transduction system.
Acta Crystallogr.,Sect.F, 65, 2009
|
|
1OMO
 
 | | alanine dehydrogenase dimer w/bound NAD (archaeal) | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SODIUM ION, alanine dehydrogenase | | Authors: | Gallagher, D.T, Smith, N.N, Holden, M.J, Schroeder, I, Monbouquette, H.G. | | Deposit date: | 2003-02-25 | | Release date: | 2004-07-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Structure of alanine dehydrogenase from Archaeoglobus: active site analysis and relation to bacterial cyclodeaminases and mammalian mu crystallin.
J.Mol.Biol., 342, 2004
|
|
5C0V
 
 | | Structure of the LARP1-unique domain DM15 | | Descriptor: | La-related protein 1, SULFATE ION | | Authors: | Lahr, R.M, Berman, A.J. | | Deposit date: | 2015-06-12 | | Release date: | 2015-08-05 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The La-related protein 1-specific domain repurposes HEAT-like repeats to directly bind a 5'TOP sequence.
Nucleic Acids Res., 43, 2015
|
|
1Y0O
 
 | |
2GCL
 
 | | Structure of the Pob3 Middle domain | | Descriptor: | CHLORIDE ION, Hypothetical 63.0 kDa protein in DAK1-ORC1 intergenic region | | Authors: | VanDemark, A.P. | | Deposit date: | 2006-03-14 | | Release date: | 2006-05-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | The Structure of the yFACT Pob3-M Domain, Its Interaction with the DNA Replication Factor RPA, and a Potential Role in Nucleosome Deposition.
Mol.Cell, 22, 2006
|
|
2GCJ
 
 | | Crystal Structure of the Pob3 middle domain | | Descriptor: | Hypothetical 63.0 kDa protein in DAK1-ORC1 intergenic region | | Authors: | VanDemark, A.P. | | Deposit date: | 2006-03-14 | | Release date: | 2006-05-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | The Structure of the yFACT Pob3-M Domain, Its Interaction with the DNA Replication Factor RPA, and a Potential Role in Nucleosome Deposition.
Mol.Cell, 22, 2006
|
|
2ECS
 
 | | Lambda Cro mutant Q27P/A29S/K32Q at 1.4 A in space group C2 | | Descriptor: | ACETATE ION, CHLORIDE ION, LITHIUM ION, ... | | Authors: | Hall, B.M, Roberts, S.A, Cordes, M.H. | | Deposit date: | 2007-02-14 | | Release date: | 2008-01-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Two structures of a lambda Cro variant highlight dimer flexibility but disfavor major dimer distortions upon specific binding of cognate DNA.
J.Mol.Biol., 375, 2008
|
|
3D8N
 
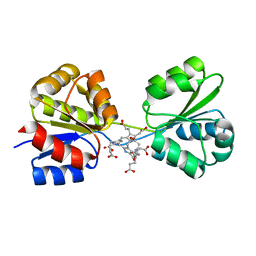 | | Uroporphyrinogen III Synthase-Uroporphyringen III Complex | | Descriptor: | 3,3',3'',3'''-[3,8,13,17-tetrakis(carboxymethyl)porphyrin-2,7,12,18-tetrayl]tetrapropanoic acid, Uroporphyrinogen-III synthase | | Authors: | Schubert, H.L. | | Deposit date: | 2008-05-23 | | Release date: | 2008-08-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and mechanistic implications of a uroporphyrinogen III synthase-product complex.
Biochemistry, 47, 2008
|
|
3D8R
 
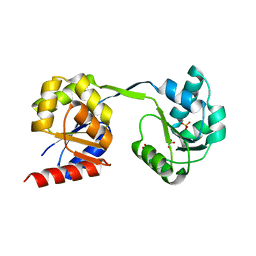 | | Thermus thermophilus Uroporphyrinogen III Synthase | | Descriptor: | PHOSPHATE ION, Uroporphyrinogen-III synthase | | Authors: | Schubert, H.L. | | Deposit date: | 2008-05-23 | | Release date: | 2008-08-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and mechanistic implications of a uroporphyrinogen III synthase-product complex.
Biochemistry, 47, 2008
|
|
3K10
 
 | |
3JRM
 
 | |
4QYZ
 
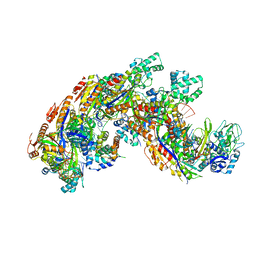 | | Crystal structure of a CRISPR RNA-guided surveillance complex, Cascade, bound to a ssDNA target | | Descriptor: | CRISPR system Cascade subunit CasA, CRISPR system Cascade subunit CasB, CRISPR system Cascade subunit CasC, ... | | Authors: | Mulepati, S, Bailey, S. | | Deposit date: | 2014-07-26 | | Release date: | 2014-09-03 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.0303 Å) | | Cite: | Structural biology. Crystal structure of a CRISPR RNA-guided surveillance complex bound to a ssDNA target.
Science, 345, 2014
|
|
