2MJY
 
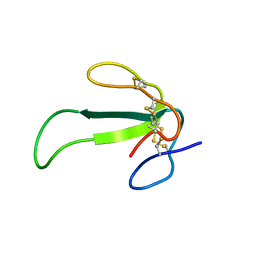 | | Solution structure of synthetic Mamba-1 peptide | | Descriptor: | Mambalgin-1 | | Authors: | He, Y, Wu, F, Tian, C. | | Deposit date: | 2014-01-21 | | Release date: | 2015-01-28 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | One-pot hydrazide-based native chemical ligation for efficient chemical synthesis and structure determination of toxin Mambalgin-1
Chem.Commun.(Camb.), 50, 2014
|
|
7LSY
 
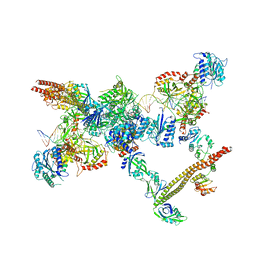 | | NHEJ Short-range synaptic complex | | Descriptor: | DNA (26-MER), DNA (5'-D(P*CP*AP*AP*TP*GP*AP*AP*AP*CP*GP*GP*AP*AP*CP*AP*GP*TP*CP*AP*G)-3'), DNA (5'-D(P*GP*TP*TP*CP*TP*TP*AP*GP*TP*AP*TP*AP*TP*A)-3'), ... | | Authors: | He, Y, Chen, S. | | Deposit date: | 2021-02-18 | | Release date: | 2021-04-14 | | Last modified: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (8.4 Å) | | Cite: | Structural basis of long-range to short-range synaptic transition in NHEJ.
Nature, 593, 2021
|
|
7LT3
 
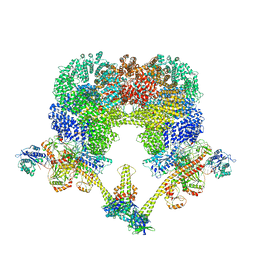 | | NHEJ Long-range synaptic complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA (30-MER), DNA (31-MER), ... | | Authors: | He, Y, Chen, S. | | Deposit date: | 2021-02-18 | | Release date: | 2021-04-14 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Structural basis of long-range to short-range synaptic transition in NHEJ.
Nature, 593, 2021
|
|
8IYS
 
 | | TUG891-bound FFAR4 in complex with Gq | | Descriptor: | 3-{4-[(4-fluoro-4'-methyl[1,1'-biphenyl]-2-yl)methoxy]phenyl}propanoic acid, Free fatty acid receptor 4, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | He, Y, Yin, H. | | Deposit date: | 2023-04-06 | | Release date: | 2023-06-21 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | Structural basis of omega-3 fatty acid receptor FFAR4 activation and G protein coupling selectivity.
Cell Res., 33, 2023
|
|
8GRR
 
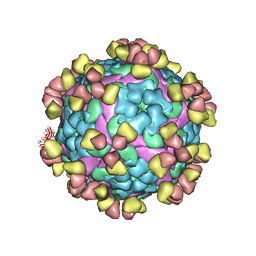 | | Complex of FMDV A/WH/CHA/09 and bovine neutralizing scFv antibody W125 | | Descriptor: | A/WH/CHA/09 VP1, A/WH/CHA/09 VP2, A/WH/CHA/09 VP3, ... | | Authors: | He, Y, Kun, L. | | Deposit date: | 2022-09-02 | | Release date: | 2023-10-11 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.72 Å) | | Cite: | Conserved antigen structures and antibody-driven variations on foot-and-mouth disease virus serotype A revealed by bovine neutralizing monoclonal antibodies.
Plos Pathog., 19, 2023
|
|
8GSP
 
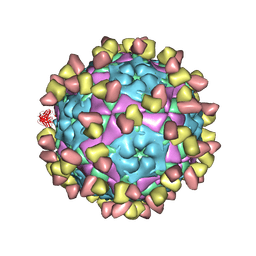 | | Complex of FMDV A/WH/CHA/09 and bovine neutralizing scFv antibody W2 | | Descriptor: | A/WH/CHA/09 VP1, A/WH/CHA/09 VP2, A/WH/CHA/09 VP3, ... | | Authors: | He, Y, Li, K. | | Deposit date: | 2022-09-06 | | Release date: | 2023-10-11 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.75 Å) | | Cite: | Conserved antigen structures and antibody-driven variations on foot-and-mouth disease virus serotype A revealed by bovine neutralizing monoclonal antibodies.
Plos Pathog., 19, 2023
|
|
4J51
 
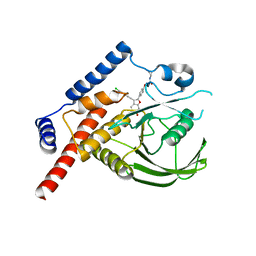 | |
7WF7
 
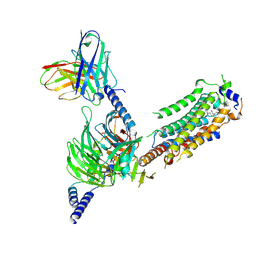 | | Cryo-EM of Sphingosine 1-phosphate receptor 1 / Gi complex bound to S1P | | Descriptor: | (2S,3R,4E)-2-amino-3-hydroxyoctadec-4-en-1-yl dihydrogen phosphate, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | He, Y, Xu, Z. | | Deposit date: | 2021-12-26 | | Release date: | 2022-01-05 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis of sphingosine-1-phosphate receptor 1 activation and biased agonism.
Nat.Chem.Biol., 18, 2022
|
|
8ZX5
 
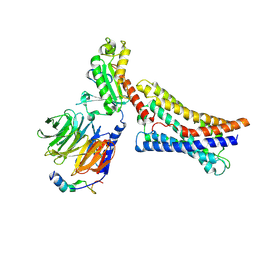 | | AM251-bound GPR55 in complex with G13 | | Descriptor: | 1-(2,4-dichlorophenyl)-5-(4-iodophenyl)-4-methyl-~{N}-piperidin-1-yl-pyrazole-3-carboxamide, G-protein coupled receptor 55, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | He, Y, Xia, R. | | Deposit date: | 2024-06-13 | | Release date: | 2024-11-13 | | Last modified: | 2025-01-22 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | Structural insight into GPR55 ligand recognition and G-protein coupling.
Cell Res., 35, 2025
|
|
8ZX4
 
 | | LPI-bound GPR55 in complex with G13 | | Descriptor: | G-protein coupled receptor 55, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | He, Y, Xia, R. | | Deposit date: | 2024-06-13 | | Release date: | 2024-11-13 | | Last modified: | 2025-01-22 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Structural insight into GPR55 ligand recognition and G-protein coupling.
Cell Res., 35, 2025
|
|
7EO2
 
 | | Cryo-EM of Sphingosine 1-phosphate receptor 1 / Gi complex bound to FTY720p | | Descriptor: | (2~{S})-2-azanyl-4-(4-octylphenyl)-2-[[oxidanyl-bis(oxidanylidene)-$l^{6}-phosphanyl]oxymethyl]butan-1-ol, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | He, Y, Xu, Z, Ikuta, T. | | Deposit date: | 2021-04-21 | | Release date: | 2022-01-05 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Structural basis of sphingosine-1-phosphate receptor 1 activation and biased agonism.
Nat.Chem.Biol., 18, 2022
|
|
7EO4
 
 | | Cryo-EM of Sphingosine 1-phosphate receptor 1 / Gi complex bound to BAF312 | | Descriptor: | 1-[[4-[(~{E})-~{N}-[[4-cyclohexyl-3-(trifluoromethyl)phenyl]methoxy]-~{C}-methyl-carbonimidoyl]-2-ethyl-phenyl]methyl]azetidine-3-carboxylic acid, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | He, Y, Xu, Z, Ikuta, T, Inoue, A. | | Deposit date: | 2021-04-21 | | Release date: | 2022-01-05 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.86 Å) | | Cite: | Structural basis of sphingosine-1-phosphate receptor 1 activation and biased agonism.
Nat.Chem.Biol., 18, 2022
|
|
6LG2
 
 | | VanR bound to Vanillate | | Descriptor: | 4-HYDROXY-3-METHOXYBENZOATE, Predicted transcriptional regulators | | Authors: | He, Y, Bharath, S.R, Song, H. | | Deposit date: | 2019-12-04 | | Release date: | 2020-02-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Developing a highly efficient hydroxytyrosol whole-cell catalyst by de-bottlenecking rate-limiting steps.
Nat Commun, 11, 2020
|
|
7EO0
 
 | | FOOT AND MOUTH DISEASE VIRUS O/TIBET/99-BOUND THE SINGLE CHAIN FRAGMEN ANTIBODY C4 | | Descriptor: | Ig heavy chain variable region, Ig lamda chain variable region, O/TIBET/99 VP1, ... | | Authors: | He, Y, Li, K. | | Deposit date: | 2021-04-21 | | Release date: | 2021-08-18 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.75 Å) | | Cite: | Two Cross-Protective Antigen Sites on Foot-and-Mouth Disease Virus Serotype O Structurally Revealed by Broadly Neutralizing Antibodies from Cattle.
J.Virol., 95, 2021
|
|
7DFL
 
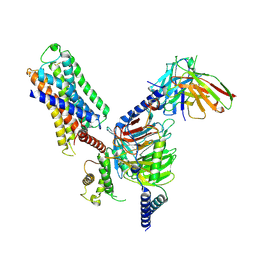 | | Cryo-EM structure of histamine H1 receptor Gq complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(q) subunit alpha, ... | | Authors: | He, Y, Xia, R, Wang, N, Xu, Z. | | Deposit date: | 2020-11-09 | | Release date: | 2021-03-31 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structure of the human histamine H 1 receptor/G q complex.
Nat Commun, 12, 2021
|
|
8BDP
 
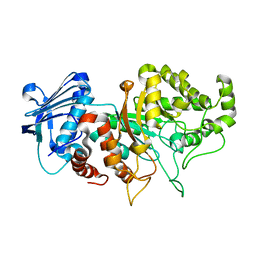 | |
8HNA
 
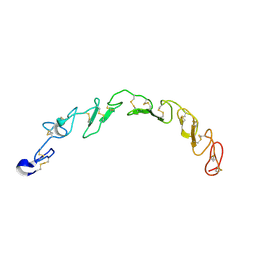 | |
8HN0
 
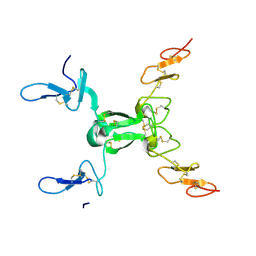 | |
5Y14
 
 | | Crystal structure of LP-40/N44 | | Descriptor: | LP-40, N44 | | Authors: | Zhang, X, Wang, X, He, Y. | | Deposit date: | 2017-07-19 | | Release date: | 2017-11-08 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.762 Å) | | Cite: | Enfuvirtide (T20)-Based Lipopeptide Is a Potent HIV-1 Cell Fusion Inhibitor: Implications for Viral Entry and Inhibition
J. Virol., 91, 2017
|
|
5XX0
 
 | |
5XWX
 
 | |
9MXZ
 
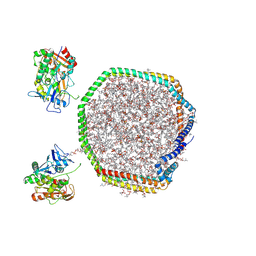 | | Lecithin:Cholesterol Acyltransferase Bound to Apolipoprotein A-I dimer in HDL | | Descriptor: | (4S,7R)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSAN-1-AMINIUM 4-OXIDE, Apolipoprotein A-I, Phosphatidylcholine-sterol acyltransferase | | Authors: | Coleman, B, Bedi, S, Hill, J.H, Morris, J, Manthei, K.A, Hart, R.C, He, Y, Shah, A.S, Jerome, W.G, Vaisar, T, Bornfeldt, K.E, Song, H, Segrest, J.P, Heinecke, J.W, Aller, S.G, Tesmer, J.J.G, Davidson, S. | | Deposit date: | 2025-01-21 | | Release date: | 2025-04-02 | | Last modified: | 2025-05-07 | | Method: | ELECTRON MICROSCOPY (9.8 Å) | | Cite: | Lecithin:cholesterol acyltransferase binds a discontinuous binding site on adjacent apolipoprotein A-I belts in HDL.
J.Lipid Res., 66, 2025
|
|
4UR9
 
 | | Structure of ligand bound glycosylhydrolase | | Descriptor: | 4-ethoxyquinazoline, CALCIUM ION, O-(2-ACETAMIDO-2-DEOXY D-GLUCOPYRANOSYLIDENE) AMINO-N-PHENYLCARBAMATE, ... | | Authors: | Darby, J.F, Landstroem, J, Roth, C, He, Y, Schultz, M, Davies, G.J, Hubbard, R.E. | | Deposit date: | 2014-06-27 | | Release date: | 2015-02-25 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of Selective Small-Molecule Activators of a Bacterial Glycoside Hydrolase.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
3ETL
 
 | | RadA recombinase from Methanococcus maripaludis in complex with AMPPNP | | Descriptor: | DNA repair and recombination protein radA, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Li, Y, He, Y, Luo, Y. | | Deposit date: | 2008-10-08 | | Release date: | 2009-05-05 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Conservation of a conformational switch in RadA recombinase from Methanococcus maripaludis.
Acta Crystallogr.,Sect.D, 65, 2009
|
|
3EWA
 
 | | RADA recombinase from METHANOCOCCUS MARIPALUDIS in complex with AMPPNP and ammonium ions | | Descriptor: | DNA repair and recombination protein radA, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Li, Y, He, Y, Luo, Y. | | Deposit date: | 2008-10-14 | | Release date: | 2009-05-05 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Conservation of a conformational switch in RadA recombinase from Methanococcus maripaludis.
Acta Crystallogr.,Sect.D, 65, 2009
|
|
