6KZ6
 
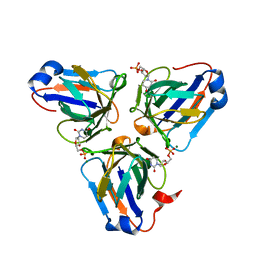 | | Crystal structure of ASFV dUTPase | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, E165R, MAGNESIUM ION | | Authors: | Guo, Y, Chen, C, Li, G.B, Cao, L, Wang, C.W. | | Deposit date: | 2019-09-23 | | Release date: | 2019-11-13 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.187 Å) | | Cite: | Structural Insight into African Swine Fever Virus dUTPase Reveals a Novel Folding Pattern in the dUTPase Family.
J.Virol., 94, 2020
|
|
1M58
 
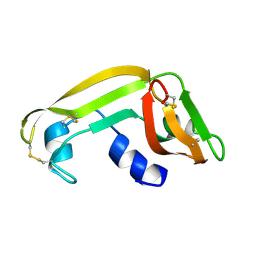 | | Solution Structure of Cytotoxic RC-RNase2 | | Descriptor: | RC-RNase2 ribonuclease | | Authors: | Hsu, C.-H, Liao, Y.-D, Wu, S.-H, Chen, C. | | Deposit date: | 2002-07-09 | | Release date: | 2003-01-09 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | 1H, 15N and 13C resonance assignments and secondary structure determination of the RC-RNase 2 from oocytes of bullfrog Rana catesbeiana.
J.Biomol.Nmr, 19, 2001
|
|
1Z5F
 
 | | Solution Structure of the Cytotoxic RC-RNase 3 with a Pyroglutamate Residue at the N-terminus | | Descriptor: | RC-RNase 3 | | Authors: | Lou, Y.C, Huang, Y.C, Pan, Y.R, Chen, C, Liao, Y.D. | | Deposit date: | 2005-03-18 | | Release date: | 2006-02-28 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Roles of N-terminal pyroglutamate in maintaining structural integrity and pKa values of catalytic histidine residues in bullfrog ribonuclease 3
J.Mol.Biol., 355, 2006
|
|
6LJB
 
 | |
6LJ9
 
 | |
6KOR
 
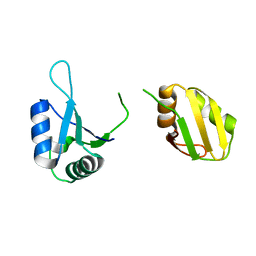 | | Crystal structure of the RRM domain of SYNCRIP | | Descriptor: | Heterogeneous nuclear ribonucleoprotein Q | | Authors: | Chen, Y, Chan, J, Chen, W, Jobichen, C. | | Deposit date: | 2019-08-12 | | Release date: | 2020-01-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.602 Å) | | Cite: | SYNCRIP, a new player in pri-let-7a processing.
Rna, 26, 2020
|
|
5BQX
 
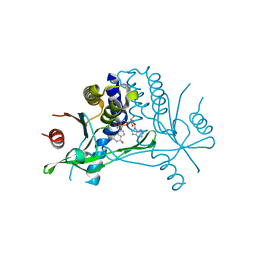 | | Crystal structure of human STING in complex with 3'2'-cGAMP | | Descriptor: | 3'2'-cGAMP, Stimulator of interferon genes protein | | Authors: | Wu, J, Zhang, X, Chen, Z.J, Chen, C. | | Deposit date: | 2015-05-29 | | Release date: | 2015-06-24 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular basis for the specific recognition of the metazoan cyclic GMP-AMP by the innate immune adaptor protein STING.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5X8Y
 
 | | A Mutation identified in Neonatal Microcephaly Destabilizes Zika Virus NS1 Assembly in vitro | | Descriptor: | ZIKV NS1 | | Authors: | Wang, D, Chen, C, Liu, S, Zhou, H, Yang, K, Zhao, Q, Ji, X, Chen, C, Xie, W, Wang, Z, Mi, L.Z, Yang, H. | | Deposit date: | 2017-03-03 | | Release date: | 2017-05-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.817 Å) | | Cite: | A Mutation Identified in Neonatal Microcephaly Destabilizes Zika Virus NS1 Assembly in Vitro
Sci Rep, 7, 2017
|
|
2LJV
 
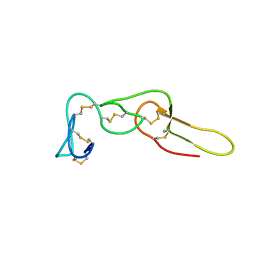 | | Solution structure of Rhodostomin G50L mutant | | Descriptor: | Disintegrin rhodostomin | | Authors: | Chuang, W, Shiu, J, Chen, C, Chen, Y, Chang, Y, Huang, C. | | Deposit date: | 2011-09-29 | | Release date: | 2012-10-03 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | Design of Integrin AlphaVbeta3-Specific Disintegrin for Cancer Therapy
To be Published
|
|
2HQ1
 
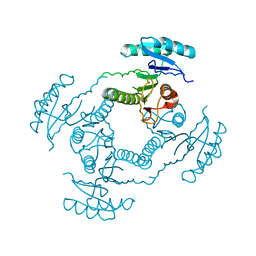 | | Crystal Structure of ORF 1438 a putative Glucose/ribitol dehydrogenase from Clostridium thermocellum | | Descriptor: | Glucose/ribitol dehydrogenase | | Authors: | Southeast Collaboratory for Structural Genomics (SECSG), Li, Y, Shaw, N, Xu, H, Cheng, C, Chen, L, Liu, Z.J, Rose, J.P, Wang, B.C. | | Deposit date: | 2006-07-18 | | Release date: | 2006-09-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of ORF 1438 a putative Glucose/ ribitol dehydrogenase from Clostridium thermocellum
To be Published
|
|
6LXN
 
 | |
6LXP
 
 | | TvCyP2 in apo form 2 | | Descriptor: | Peptidyl-prolyl cis-trans isomerase | | Authors: | Aryal, S, Chen, C, Hsu, C.H. | | Deposit date: | 2020-02-11 | | Release date: | 2020-09-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | N-Terminal Segment of TvCyP2 Cyclophilin fromTrichomonas vaginalisIs Involved in Self-Association, Membrane Interaction, and Subcellular Localization.
Biomolecules, 10, 2020
|
|
1ZKZ
 
 | | Crystal Structure of BMP9 | | Descriptor: | Growth/differentiation factor 2 | | Authors: | Brown, M.A, Zhao, Q, Baker, K.A, Naik, C, Chen, C, Pukac, L, Singh, M, Tsareva, T, Parice, Y, Mahoney, A, Roschke, V, Sanyal, I, Choe, S. | | Deposit date: | 2005-05-04 | | Release date: | 2005-05-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Crystal structure of BMP-9 and functional interactions with pro-region and receptors
J.Biol.Chem., 280, 2005
|
|
1X37
 
 | | Structure of Bacillus subtilis Lon protease SSD domain | | Descriptor: | ATP-dependent protease La 1 | | Authors: | Wang, I, Lou, Y.C, Lo, S.C, Lee, Y.L, Wu, S.H, Chen, C. | | Deposit date: | 2005-04-30 | | Release date: | 2005-10-30 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural basis and DNA binding property of SSD domain of Bacillus subtilis Lon protease
to be published
|
|
3EWP
 
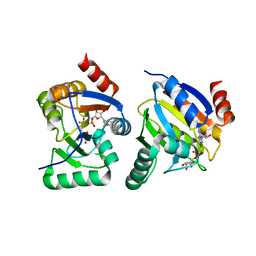 | | complex of substrate ADP-ribose with IBV Nsp3 ADRP domain | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, Non-structural protein 3 | | Authors: | Xu, Y, Cong, L, Chen, C, Wei, L, Zhao, Q, Xu, X, Ma, Y, Bartlam, M, Rao, Z. | | Deposit date: | 2008-10-16 | | Release date: | 2009-01-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of two coronavirus ADP-ribose-1''-monophosphatases and their complexes with ADP-Ribose: a systematic structural analysis of the viral ADRP domain.
J.Virol., 83, 2009
|
|
3EWQ
 
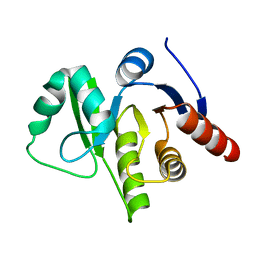 | | HCov-229E Nsp3 ADRP domain | | Descriptor: | Non-structural protein 3 | | Authors: | Xu, Y, Cong, L, Chen, C, Wei, L, Zhao, Q, Xu, X, Ma, Y, Bartlam, M, Rao, Z. | | Deposit date: | 2008-10-16 | | Release date: | 2009-01-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of two coronavirus ADP-ribose-1''-monophosphatases and their complexes with ADP-Ribose: a systematic structural analysis of the viral ADRP domain.
J.Virol., 83, 2009
|
|
3EWR
 
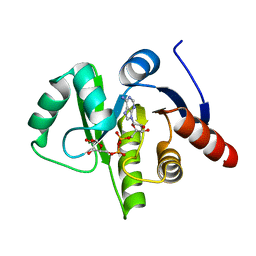 | | complex of substrate ADP-ribose with HCoV-229E Nsp3 ADRP domain | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, Non-structural protein 3 | | Authors: | Xu, Y, Cong, L, Chen, C, Wei, L, Zhao, Q, Xu, X, Ma, Y, Bartlam, M, Rao, Z. | | Deposit date: | 2008-10-16 | | Release date: | 2009-01-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Crystal structures of two coronavirus ADP-ribose-1''-monophosphatases and their complexes with ADP-Ribose: a systematic structural analysis of the viral ADRP domain.
J.Virol., 83, 2009
|
|
3EWO
 
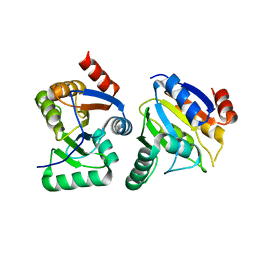 | | IBV Nsp3 ADRP domain | | Descriptor: | Non-structural protein 3 | | Authors: | Xu, Y, Cong, L, Chen, C, Wei, L, Zhao, Q, Xu, X, Ma, Y, Bartlam, M, Rao, Z. | | Deposit date: | 2008-10-16 | | Release date: | 2009-01-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of two coronavirus ADP-ribose-1''-monophosphatases and their complexes with ADP-Ribose: a systematic structural analysis of the viral ADRP domain.
J.Virol., 83, 2009
|
|
1JLZ
 
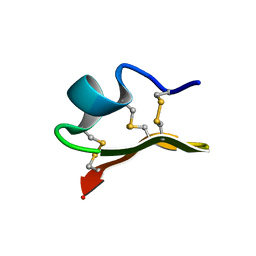 | | Solution Structure of a K+-Channel Blocker from the Scorpion Toxin of Tityus cambridgei | | Descriptor: | Tityustoxin alpha-KTx | | Authors: | Wang, I, Wu, S.-H, Chang, H.-K, Shieh, R.-C, Yu, H.-M, Chen, C. | | Deposit date: | 2001-07-17 | | Release date: | 2002-02-06 | | Last modified: | 2024-10-09 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a K(+)-channel blocker from the scorpion Tityus cambridgei.
Protein Sci., 11, 2002
|
|
3FUQ
 
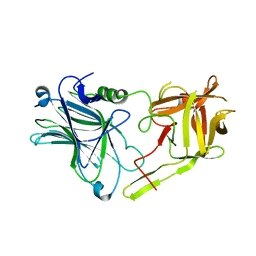 | | Glycosylated SV2 and Gangliosides as Dual Receptors for Botulinum Neurotoxin Serotype F | | Descriptor: | BoNT/F (Neurotoxin type F) | | Authors: | Fu, Z, Chen, C, Barbieri, J.T, Kim, J.-J.P, Baldwin, M.R. | | Deposit date: | 2009-01-14 | | Release date: | 2009-06-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Glycosylated SV2 and gangliosides as dual receptors for botulinum neurotoxin serotype F
Biochemistry, 48, 2009
|
|
3FUO
 
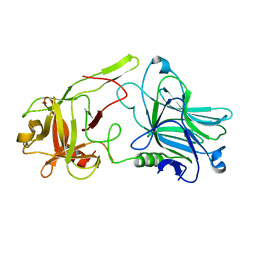 | | The Crystal structure of receptor binding domain of botulinum neurotoxin serotype A | | Descriptor: | Botulinum neurotoxin type A | | Authors: | Fu, Z, Chen, C, Barbieri, J.T, Kim, J.-J.P, Baldwin, M.R. | | Deposit date: | 2009-01-14 | | Release date: | 2009-06-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Glycosylated SV2 and gangliosides as dual receptors for botulinum neurotoxin serotype F
Biochemistry, 48, 2009
|
|
1XHH
 
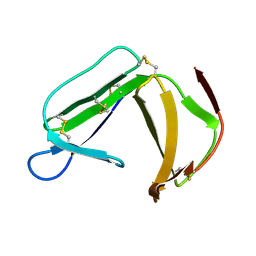 | | Solution Structure of porcine beta-microseminoprotein | | Descriptor: | beta-microseminoprotein | | Authors: | Wang, I, Lou, Y.C, Wu, K.P, Wu, S.H, Chang, W.C, Chen, C. | | Deposit date: | 2004-09-20 | | Release date: | 2005-03-20 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | Novel solution structure of porcine beta-microseminoprotein
J.Mol.Biol., 346, 2005
|
|
1EDP
 
 | |
4B27
 
 | | Trp RNA-binding attenuation protein: modifying symmetry and stability of a circular oligomer | | Descriptor: | TRANSCRIPTION ATTENUATION PROTEIN MTRB, TRYPTOPHAN | | Authors: | Bayfield, O.W, Chen, C, Patterson, A.R, Luan, W, Smits, C, Gollnick, P, Antson, A.A. | | Deposit date: | 2012-07-12 | | Release date: | 2012-09-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Trp RNA-Binding Attenuation Protein: Modifying Symmetry and Stability of a Circular Oligomer.
Plos One, 7, 2012
|
|
4TOR
 
 | | Crystal structure of Tankyrase 1 with IWR-8 | | Descriptor: | 1-[(1-acetyl-5-bromo-1H-indol-6-yl)sulfonyl]-N-ethyl-N-(3-methylphenyl)piperidine-4-carboxamide, CHLORIDE ION, Tankyrase-1, ... | | Authors: | Chen, H, Zhang, X, Lum, L, Chen, C. | | Deposit date: | 2014-06-06 | | Release date: | 2015-05-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.501 Å) | | Cite: | Disruption of Wnt/ beta-Catenin Signaling and Telomeric Shortening Are Inextricable Consequences of Tankyrase Inhibition in Human Cells.
Mol.Cell.Biol., 35, 2015
|
|
