6D4L
 
 | |
3KCL
 
 | |
3KBJ
 
 | |
3KCO
 
 | |
3KBN
 
 | |
3KYU
 
 | |
3KYV
 
 | |
3KYW
 
 | |
3KYX
 
 | |
6FJI
 
 | |
6FJJ
 
 | |
6D54
 
 | |
6GCY
 
 | | Joint neutron and x-ray crystal structure of human carbonic anhydrase IX mimic (saccharin-sugar conjugate complex) | | Descriptor: | Carbonic anhydrase 2, ZINC ION, [1-(1,1-dioxido-3-oxo-2,3-dihydro-1,2-benzothiazol-6-yl)-1H-1,2,3-triazol-4-yl]methyl alpha-L-idopyranoside | | Authors: | Fisher, S.Z, Koruza, K. | | Deposit date: | 2018-04-20 | | Release date: | 2019-02-06 | | Last modified: | 2024-05-01 | | Method: | NEUTRON DIFFRACTION (1.3 Å), X-RAY DIFFRACTION | | Cite: | Using neutron crystallography to elucidate the basis of selective inhibition of carbonic anhydrase by saccharin and a derivative.
J. Struct. Biol., 205, 2019
|
|
3KBS
 
 | |
3KBM
 
 | |
3KBV
 
 | |
3KCJ
 
 | |
3KBW
 
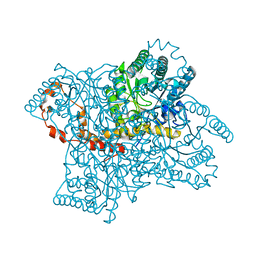 | |
6E21
 
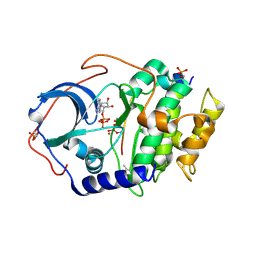 | | Joint X-ray/neutron structure of PKAc with products Sr2-ADP and phosphorylated peptide SP20 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, STRONTIUM ION, cAMP-dependent protein kinase catalytic subunit alpha, ... | | Authors: | Kovalevsky, A, Gerlits, O.O, Taylor, S. | | Deposit date: | 2018-07-10 | | Release date: | 2019-04-03 | | Last modified: | 2023-10-25 | | Method: | NEUTRON DIFFRACTION (2 Å), X-RAY DIFFRACTION | | Cite: | Zooming in on protons: Neutron structure of protein kinase A trapped in a product complex.
Sci Adv, 5, 2019
|
|
6GTL
 
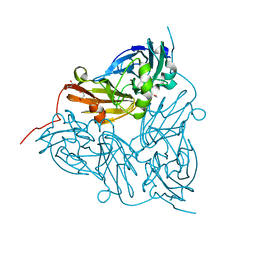 | | Achromobacter cycloclastes copper nitrite reductase at pH 6.0 | | Descriptor: | COPPER (II) ION, Copper-containing nitrite reductase, MALONATE ION, ... | | Authors: | Halsted, T.P, Eady, R.R, Antonyuk, S.V, Hasnain, S.S. | | Deposit date: | 2018-06-18 | | Release date: | 2019-07-03 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Catalytically important damage-free structures of a copper nitrite reductase obtained by femtosecond X-ray laser and room-temperature neutron crystallography.
Iucrj, 6, 2019
|
|
3KYY
 
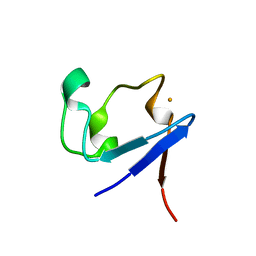 | |
4JEC
 
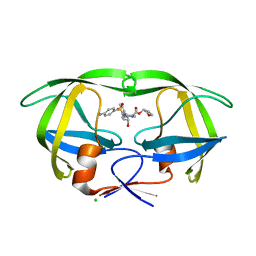 | | Joint neutron and X-ray structure of per-deuterated HIV-1 protease in complex with clinical inhibitor amprenavir | | Descriptor: | CHLORIDE ION, HIV-1 protease, {3-[(4-AMINO-BENZENESULFONYL)-ISOBUTYL-AMINO]-1-BENZYL-2-HYDROXY-PROPYL}-CARBAMIC ACID TETRAHYDRO-FURAN-3-YL ESTER | | Authors: | Kovalevsky, A.Y, Weber, I.T, Langan, P. | | Deposit date: | 2013-02-26 | | Release date: | 2013-07-24 | | Last modified: | 2024-02-28 | | Method: | NEUTRON DIFFRACTION (2.01 Å), X-RAY DIFFRACTION | | Cite: | Joint X-ray/Neutron Crystallographic Study of HIV-1 Protease with Clinical Inhibitor Amprenavir: Insights for Drug Design.
J.Med.Chem., 56, 2013
|
|
4DVO
 
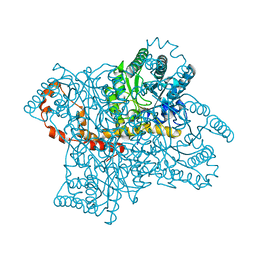 | | Room-temperature joint X-ray/neutron structure of D-xylose isomerase in complex with 2Ni2+ and per-deuterated D-sorbitol at pH 5.9 | | Descriptor: | NICKEL (II) ION, Xylose isomerase, sorbitol | | Authors: | Kovalevsky, A.Y, Hanson, L, Langan, P. | | Deposit date: | 2012-02-23 | | Release date: | 2012-08-29 | | Last modified: | 2024-02-28 | | Method: | NEUTRON DIFFRACTION (2 Å), X-RAY DIFFRACTION | | Cite: | Inhibition of D-xylose isomerase by polyols: atomic details by joint X-ray/neutron crystallography.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4DUO
 
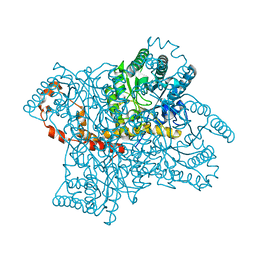 | | Room-temperature X-ray structure of D-Xylose Isomerase in complex with 2Mg2+ ions and xylitol at pH 7.7 | | Descriptor: | MAGNESIUM ION, Xylitol, Xylose isomerase | | Authors: | Kovalevsky, A.Y, Hanson, L, Langan, P. | | Deposit date: | 2012-02-22 | | Release date: | 2012-08-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Inhibition of D-xylose isomerase by polyols: atomic details by joint X-ray/neutron crystallography.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
5T8H
 
 | |
