5WCA
 
 | | Crystal structure of the broadly neutralizing Influenza A antibody VRC 315 27-1C08 Fab. | | Descriptor: | VRC315 27-1C08 Fab Heavy chain, VRC315 27-1C08 Fab Light chain | | Authors: | Joyce, M.G, Andrews, S.F, Mascola, J.R, McDermott, A.B, Kwong, P.D. | | Deposit date: | 2017-06-29 | | Release date: | 2017-08-02 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.369 Å) | | Cite: | Preferential induction of cross-group influenza A hemagglutinin stem-specific memory B cells after H7N9 immunization in humans.
Sci Immunol, 2, 2017
|
|
5WCD
 
 | | Crystal structure of the broadly neutralizing Influenza A antibody VRC 315 04-1D02 Fab. | | Descriptor: | PHOSPHATE ION, SULFATE ION, VRC315 04-1D02 Fab Heavy chain, ... | | Authors: | Joyce, M.G, Andrews, S.F, Mascola, J.R, McDermott, A.B, Kwong, P.D. | | Deposit date: | 2017-06-29 | | Release date: | 2017-08-23 | | Last modified: | 2018-11-28 | | Method: | X-RAY DIFFRACTION (1.814 Å) | | Cite: | Preferential induction of cross-group influenza A hemagglutinin stem-specific memory B cells after H7N9 immunization in humans.
Sci Immunol, 2, 2017
|
|
5ZBU
 
 | |
5YWR
 
 | | Crystal Structure of RING E3 ligase ZNRF1 in complex with Ube2N (Ubc13) | | Descriptor: | E3 ubiquitin-protein ligase ZNRF1, FORMIC ACID, TRIETHYLENE GLYCOL, ... | | Authors: | Behera, A.P, Naskar, P, Datta, A.B. | | Deposit date: | 2017-11-30 | | Release date: | 2018-06-06 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Structural insights into the nanomolar affinity of RING E3 ligase ZNRF1 for Ube2N and its functional implications.
Biochem. J., 475, 2018
|
|
5W9H
 
 | |
5W9I
 
 | |
5WGP
 
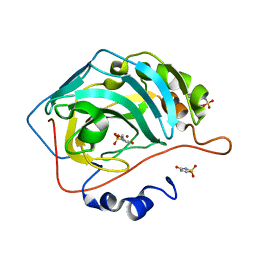 | | Human Carbonic Anhydrase IX-mimic complexed with AceK | | Descriptor: | Acesulfame, Carbonic anhydrase 2, ZINC ION | | Authors: | Murray, A.B, Lomelino, C.L, Supuran, C.T, McKenna, R. | | Deposit date: | 2017-07-14 | | Release date: | 2018-02-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | "Seriously Sweet": Acesulfame K Exhibits Selective Inhibition Using Alternative Binding Modes in Carbonic Anhydrase Isoforms.
J. Med. Chem., 61, 2018
|
|
5W9L
 
 | |
5ZC4
 
 | |
5W9K
 
 | |
1XVY
 
 | | Crystal Structure of iron-free Serratia marcescens SfuA | | Descriptor: | CITRIC ACID, sfuA | | Authors: | Shouldice, S.R, McRee, D.E, Dougan, D.R, Tari, L.W, Schryvers, A.B. | | Deposit date: | 2004-10-28 | | Release date: | 2004-12-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Novel Anion-independent Iron Coordination by Members of a Third Class of Bacterial Periplasmic Ferric Ion-binding Proteins
J.Biol.Chem., 280, 2005
|
|
1YK8
 
 | | Cathepsin K complexed with a cyanamide-based inhibitor | | Descriptor: | Cathepsin K, TERT-BUTYL 2-CYANO-2-METHYLHYDRAZINECARBOXYLATE | | Authors: | Barrett, D.G, Deaton, D.N, Hassell, A.M, McFadyen, R.B, Miller, A.B, Miller, L.R, Payne, J.A, Shewchuk, L.M, Willard, D.H, Wright, L.L. | | Deposit date: | 2005-01-17 | | Release date: | 2005-07-19 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Acyclic cyanamide-based inhibitors of cathepsin K.
Bioorg.Med.Chem.Lett., 15, 2005
|
|
1YK7
 
 | | Cathepsin K complexed with a cyanopyrrolidine inhibitor | | Descriptor: | Cathepsin K, N2-[(BENZYLOXY)CARBONYL]-N1-[(3S)-1-CYANOPYRROLIDIN-3-YL]-L-LEUCINAMIDE | | Authors: | Barrett, D.G, Deaton, D.N, Hassell, A.M, McFadyen, R.B, Miller, A.B, Miller, L.R, Shewchuk, L.M, Tavares, F.X, Willard, D.H, Wright, L.L. | | Deposit date: | 2005-01-17 | | Release date: | 2005-03-22 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Novel and potent cyclic cyanamide-based cathepsin K inhibitors.
Bioorg.Med.Chem.Lett., 15, 2005
|
|
1XF6
 
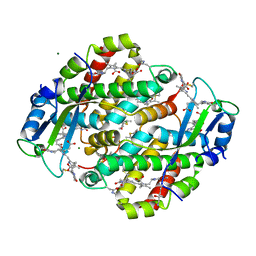 | | High resolution crystal structure of phycoerythrin 545 from the marine cryptophyte rhodomonas CS24 | | Descriptor: | 15,16-DIHYDROBILIVERDIN, B-phycoerythrin beta chain, CHLORIDE ION, ... | | Authors: | Doust, A.B, Marai, C.N.J, Harrop, S.J, Wilk, K.E, Curmi, P.M.G, Scholes, G.D. | | Deposit date: | 2004-09-14 | | Release date: | 2004-11-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Developing a structure-function model for the cryptophyte phycoerythrin 545 using ultrahigh resolution crystallography and ultrafast laser spectroscopy
J.Mol.Biol., 344, 2004
|
|
1XG0
 
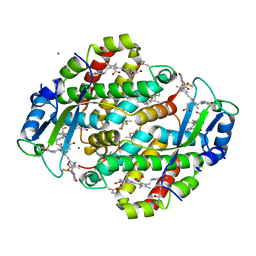 | | High resolution crystal structure of phycoerythrin 545 from the marine cryptophyte rhodomonas CS24 | | Descriptor: | 15,16-DIHYDROBILIVERDIN, B-phycoerythrin beta chain, CHLORIDE ION, ... | | Authors: | Doust, A.B, Marai, C.N.J, Harrop, S.J, Wilk, K.E, Curmi, P.M.G, Scholes, G.D. | | Deposit date: | 2004-09-16 | | Release date: | 2004-11-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | Developing a structure-function model for the cryptophyte phycoerythrin 545 using ultrahigh resolution crystallography and ultrafast laser spectroscopy
J.Mol.Biol., 344, 2004
|
|
1XVX
 
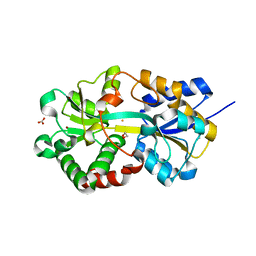 | | Crystal Structure of iron-loaded Yersinia enterocolitica YfuA | | Descriptor: | 1,2-ETHANEDIOL, CARBONATE ION, FE (III) ION, ... | | Authors: | Shouldice, S.R, McRee, D.E, Dougan, D.R, Tari, L.W, Schryvers, A.B. | | Deposit date: | 2004-10-28 | | Release date: | 2004-12-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Novel Anion-independent Iron Coordination by Members of a Third Class of Bacterial Periplasmic Ferric Ion-binding Proteins
J.Biol.Chem., 280, 2005
|
|
1ZPU
 
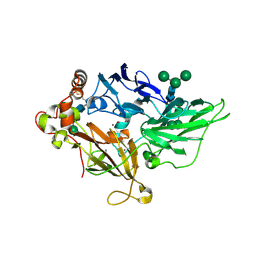 | | Crystal Structure of Fet3p, a Multicopper Oxidase that Functions in Iron Import | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, COPPER (I) ION, ... | | Authors: | Taylor, A.B, Stoj, C.S, Ziegler, L, Kosman, D.J, Hart, P.J. | | Deposit date: | 2005-05-17 | | Release date: | 2005-10-04 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The copper-iron connection in biology: Structure of the metallo-oxidase Fet3p.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
1YT7
 
 | | Cathepsin K complexed with a constrained ketoamide inhibitor | | Descriptor: | (1R)-2,2-DIMETHYL-1-({5-[4-(TRIFLUOROMETHYL)PHENYL]-1,3,4-OXADIAZOL-2-YL}METHYL)PROPYL (1S)-1-{OXO[(2-OXO-1,3-OXAZOLIDIN-3-YL)AMINO]ACETYL}PENTYLCARBAMATE, Cathepsin K, SULFATE ION | | Authors: | Barrett, D.G, Boncek, V.M, Catalano, J.G, Deaton, D.N, Hassell, A.M, Jurgensen, C.H, Long, S.T, McFadyen, R.B, Miller, A.B, Miller, L.R, Payne, J.A, Ray, J.A, Samano, V, Shewchuk, L.M, Tavares, F.X, Wells-Knecht, K.J, Willard, D.H, Wright, L.L, Zhou, H.Q. | | Deposit date: | 2005-02-10 | | Release date: | 2005-07-19 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | P(2)-P(3) conformationally constrained ketoamide-based inhibitors of cathepsin K.
Bioorg.Med.Chem.Lett., 15, 2005
|
|
1XWR
 
 | | Crystal structure of the coliphage lambda transcription activator protein CII | | Descriptor: | ISOPROPYL ALCOHOL, Regulatory protein CII | | Authors: | Datta, A.B, Panjikar, S, Weiss, M.S, Chakrabarti, P, Parrack, P. | | Deposit date: | 2004-11-02 | | Release date: | 2005-06-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Structure of {lambda} CII: Implications for recognition of direct-repeat DNA by an unusual tetrameric organization
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
1Y4L
 
 | | Crystal structure of Bothrops asper myotoxin II complexed with the anti-trypanosomal drug suramin | | Descriptor: | 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, 8,8'-[CARBONYLBIS[IMINO-3,1-PHENYLENECARBONYLIMINO(4-METHYL-3,1-PHENYLENE)CARBONYLIMINO]]BIS-1,3,5-NAPHTHALENETRISULFONIC ACID, ISOPROPYL ALCOHOL, ... | | Authors: | Murakami, M.T, Arruda, E.Z, Melo, P.A, Martinez, A.B, Calil-Elias, S, Tomaz, M.A, Lomonte, B, Gutierrez, J.M, Arni, R.K. | | Deposit date: | 2004-12-01 | | Release date: | 2005-06-28 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Inhibition of Myotoxic Activity of Bothrops asper Myotoxin II by the Anti-trypanosomal Drug Suramin.
J.Mol.Biol., 350, 2005
|
|
2AUX
 
 | | Cathepsin K complexed with a semicarbazone inhibitor | | Descriptor: | (1R)-2-METHYL-1-(PHENYLMETHYL)PROPYL[(1S)-1-FORMYLPENTYL]CARBAMATE, Cathepsin K | | Authors: | Adkison, K.K, Barrett, D.G, Deaton, D.N, Gampe, R.T, Hassell, A.M, Long, S.T, McFadyen, R.B, Miller, A.B, Miller, L.R, Shewchuk, L.M. | | Deposit date: | 2005-08-29 | | Release date: | 2006-08-08 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Semicarbazone-based inhibitors of cathepsin K, are they prodrugs for aldehyde inhibitors?
Bioorg.Med.Chem.Lett., 16, 2006
|
|
2AUZ
 
 | | Cathepsin K complexed with a semicarbazone inhibitor | | Descriptor: | 1-(PHENYLMETHYL)CYCLOPENTYL[(1S)-1-FORMYLPENTYL]CARBAMATE, Cathepsin K, SULFATE ION | | Authors: | Adkison, K.K, Barrett, D.G, Deaton, D.N, Gampe, R.T, Hassell, A.M, Long, S.T, McFadyen, R.B, Miller, A.B, Miller, L.R, Shewchuk, L.M. | | Deposit date: | 2005-08-29 | | Release date: | 2006-08-08 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Semicarbazone-based inhibitors of cathepsin K, are they prodrugs for aldehyde inhibitors?
Bioorg.Med.Chem.Lett., 16, 2006
|
|
5LA6
 
 | | Tubulin-pironetin complex | | Descriptor: | CALCIUM ION, GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Prota, A.E, Setter, J, Waight, A.B, Bargsten, K, Murga, J, Diaz, J.F, Steinmetz, M.O. | | Deposit date: | 2016-06-13 | | Release date: | 2016-07-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Pironetin Binds Covalently to alpha Cys316 and Perturbs a Major Loop and Helix of alpha-Tubulin to Inhibit Microtubule Formation.
J.Mol.Biol., 428, 2016
|
|
5LFY
 
 | |
5LME
 
 | | Specific-DNA binding activity of the cross-brace zinc finger motif of the piggyBac transposase | | Descriptor: | ZINC ION, piggyBac transposase | | Authors: | Morellet, N, Taylor, J.A, Wieninger, S, Moriau, S, Li, X, Lescop, E, Mathy, N, Bischerour, J, Betermier, M, Bardiaux, B, Nilges, M, Craig, N.L, Hickman, A.B, Dyda, F, Guittet, E. | | Deposit date: | 2016-07-30 | | Release date: | 2017-12-20 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Sequence-specific DNA binding activity of the cross-brace zinc finger motif of the piggyBac transposase.
Nucleic Acids Res., 46, 2018
|
|
