5E34
 
 | |
25C8
 
 | | CATALYTIC ANTIBODY 5C8, FAB-HAPTEN COMPLEX | | Descriptor: | IGG 5C8, N-METHYL-N-(PARA-GLUTARAMIDOPHENYL-ETHYL)-PIPERIDINIUM ION | | Authors: | Gruber, K, Wilson, I.A. | | Deposit date: | 1998-03-18 | | Release date: | 1999-03-23 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for antibody catalysis of a disfavored ring closure reaction.
Biochemistry, 38, 1999
|
|
7RDM
 
 | |
7RDJ
 
 | | Crystal structure of PCDN-16B, an anti-HIV antibody from the PCDN bnAb lineage (non-cysteinylated state) | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, PCDN-16B Fab heavy chain, ... | | Authors: | Omorodion, O, Wilson, I.A. | | Deposit date: | 2021-07-09 | | Release date: | 2021-11-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structural and Biochemical Characterization of Cysteinylation in Broadly Neutralizing Antibodies to HIV-1.
J.Mol.Biol., 433, 2021
|
|
7RDK
 
 | | Crystal structure of PCDN-16B, an anti-HIV antibody from the PCDN bnAb lineage (cysteinylated state) | | Descriptor: | 1,2-ETHANEDIOL, CYSTEINE, GLYCEROL, ... | | Authors: | Omorodion, O, Wilson, I.A. | | Deposit date: | 2021-07-09 | | Release date: | 2021-11-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Structural and Biochemical Characterization of Cysteinylation in Broadly Neutralizing Antibodies to HIV-1.
J.Mol.Biol., 433, 2021
|
|
7RDL
 
 | |
6W05
 
 | |
7RDH
 
 | |
6W00
 
 | | Crystal structure of Fab239 in complex with NPNA2 peptide from circumsporozoite protein | | Descriptor: | Fab239 heavy chain, Fab239 light chain, Immunoglobulin G-binding protein G, ... | | Authors: | Pholcharee, T, Oyen, D, Wilson, I.A. | | Deposit date: | 2020-02-28 | | Release date: | 2020-07-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.853 Å) | | Cite: | Structural and biophysical correlation of anti-NANP antibodies with in vivo protection against P. falciparum.
Nat Commun, 12, 2021
|
|
1ZHN
 
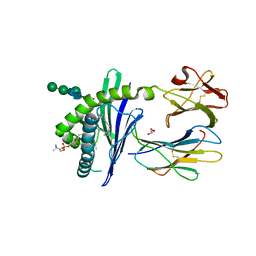 | | Crystal Structure of mouse CD1d bound to the self ligand phosphatidylcholine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 7-[(DODECANOYLOXY)METHYL]-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-3,5,8-TRIOXA-4-PHOSPHADOTRIACONTAN-1-AMINIUM 4-OXIDE, CD1d1 antigen, ... | | Authors: | Giabbai, B, Sidobre, S, Crispin, M.M.D, Sanchez Ruiz, Y, Bachi, A, Kronenberg, M, Wilson, I.A, Degano, M. | | Deposit date: | 2005-04-26 | | Release date: | 2005-07-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of mouse CD1d bound to the self ligand phosphatidylcholine: a molecular basis for NKT cell activation
J.Immunol., 175, 2005
|
|
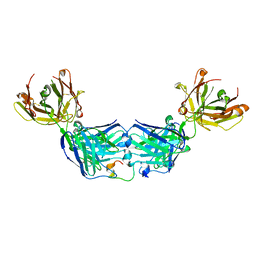
6WG1
 
 | |
2CKB
 
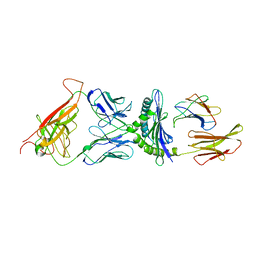 | | STRUCTURE OF THE 2C/KB/DEV8 COMPLEX | | Descriptor: | ALPHA, BETA T CELL RECEPTOR, BETA-2 MICROGLOBULIN, ... | | Authors: | Garcia, K.C, Degano, M, Wilson, I.A. | | Deposit date: | 1998-01-14 | | Release date: | 1998-04-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis of plasticity in T cell receptor recognition of a self peptide-MHC antigen.
Science, 279, 1998
|
|
5JOP
 
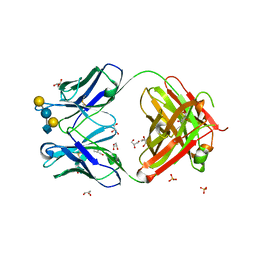 | | Crystal structure of anti-glycan antibody Fab14.22 in complex with Streptococcus pneumoniae serotype 14 tetrasaccharide at 1.75 A | | Descriptor: | Fab 14.22 light chain, Fab14.22 heavy chain, GLYCEROL, ... | | Authors: | Sarkar, A, Irimia, A, Teyton, L, Wilson, I.A. | | Deposit date: | 2016-05-02 | | Release date: | 2017-03-22 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | T cells control the generation of nanomolar-affinity anti-glycan antibodies.
J. Clin. Invest., 127, 2017
|
|
5KUX
 
 | |
5KUY
 
 | |
5IF0
 
 | |
5IDL
 
 | | Crystal structure of the germline-targeting HIV-1 gp120 engineered outer domain, eOD-GT8 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Germline-targeting HIV-1 gp120 engineered outer domain, eOD-GT8 | | Authors: | Julien, J.P, Ereno-Orbea, J, Jardine, J.G, Schief, W.R, Wilson, I.A. | | Deposit date: | 2016-02-24 | | Release date: | 2016-10-05 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | HIV-1 broadly neutralizing antibody precursor B cells revealed by germline-targeting immunogen.
Science, 351, 2016
|
|
5IES
 
 | |
5IFA
 
 | |
5ILT
 
 | | Crystal structure of bovine Fab A01 | | Descriptor: | bovine Fab A01 heavy chain, bovine Fab A01 light chain | | Authors: | Stanfield, R.L, Wilson, I.A. | | Deposit date: | 2016-03-04 | | Release date: | 2016-08-24 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Conservation and diversity in the ultralong third heavy-chain complementarity-determining region of bovine antibodies.
Sci Immunol, 1, 2016
|
|
5IJV
 
 | | Crystal structure of bovine Fab E03 | | Descriptor: | bovine Fab E03 heavy chain, bovine Fab E03 light chain | | Authors: | Stanfield, R.L, Wilson, I.A. | | Deposit date: | 2016-03-02 | | Release date: | 2016-10-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Conservation and diversity in the ultralong third heavy-chain complementarity-determining region of bovine antibodies.
Sci Immunol, 1, 2016
|
|
5HZQ
 
 | | Crystal structure of cellular retinoic acid binding protein 2 (CRABP2)-aryl fluorosulfate covalent conjugate | | Descriptor: | 4'-[(3,6,9,12-tetraoxapentadec-14-yn-1-yl)oxy][1,1'-biphenyl]-4-yl sulfurofluoridate, Cellular retinoic acid-binding protein 2, GLYCEROL | | Authors: | Chen, W, Mortenson, D.E, Wilson, I.A, Kelly, J.W. | | Deposit date: | 2016-02-02 | | Release date: | 2016-06-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Arylfluorosulfates Inactivate Intracellular Lipid Binding Protein(s) through Chemoselective SuFEx Reaction with a Binding Site Tyr Residue.
J.Am.Chem.Soc., 138, 2016
|
|
5YL9
 
 | |
5ZUV
 
 | | Crystal Structure of the Human Coronavirus 229E HR1 motif in complex with pan-CoVs inhibitor EK1 | | Descriptor: | CHLORIDE ION, Spike glycoprotein,Spike glycoprotein,inhibitor EK1 | | Authors: | Yan, L, Yang, B, Wilson, I.A. | | Deposit date: | 2018-05-08 | | Release date: | 2019-04-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | A pan-coronavirus fusion inhibitor targeting the HR1 domain of human coronavirus spike.
Sci Adv, 5, 2019
|
|
5ZVM
 
 | |
